| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
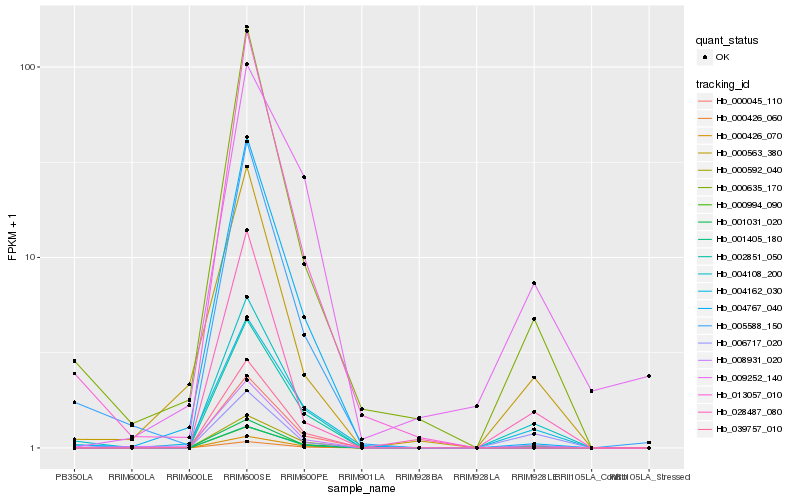
Hb_004108_200 |
0.0 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004162_030 |
0.1043955361 |
- |
- |
PREDICTED: lysosomal Pro-X carboxypeptidase-like [Jatropha curcas] |
| 3 |
Hb_001031_020 |
0.1354418027 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 4 |
Hb_000635_170 |
0.1379442678 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_000045_110 |
0.151721509 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 6 |
Hb_008931_020 |
0.1572711779 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 7 |
Hb_006717_020 |
0.1673103955 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 8 |
Hb_028487_080 |
0.1696119542 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000563_380 |
0.1769651675 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |
| 10 |
Hb_004767_040 |
0.1837134224 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 11 |
Hb_009252_140 |
0.187568159 |
- |
- |
Spotted leaf protein, putative [Ricinus communis] |
| 12 |
Hb_001405_180 |
0.1876279453 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_039757_010 |
0.1878408667 |
- |
- |
- |
| 14 |
Hb_000426_060 |
0.1881762144 |
- |
- |
hypothetical protein CISIN_1g048192mg, partial [Citrus sinensis] |
| 15 |
Hb_005588_150 |
0.1882384519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002851_050 |
0.1891109297 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_000994_090 |
0.190123677 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 18 |
Hb_013057_010 |
0.1904777894 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 19 |
Hb_000426_070 |
0.1909472484 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 20 |
Hb_000592_040 |
0.1911088139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |