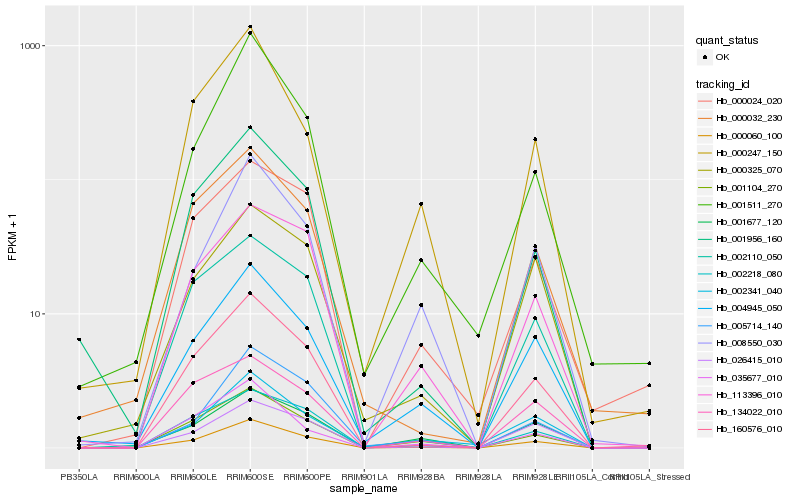
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000060_100 |
0.0 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_160576_010 |
0.0661239395 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004945_050 |
0.0937226919 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 4 |
Hb_001104_270 |
0.1032112091 |
- |
- |
hypothetical protein POPTR_0007s13980g [Populus trichocarpa] |
| 5 |
Hb_002218_080 |
0.1115282505 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 6 |
Hb_002110_050 |
0.1162130393 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001956_160 |
0.1165250194 |
- |
- |
hypothetical protein L484_009438 [Morus notabilis] |
| 8 |
Hb_008550_030 |
0.1176509089 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 9 |
Hb_026415_010 |
0.1202619347 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 10 |
Hb_113396_010 |
0.1227976072 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_001511_270 |
0.127665902 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 12 |
Hb_005714_140 |
0.128503089 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_002341_040 |
0.1286843216 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 14 |
Hb_000032_230 |
0.1301062102 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20463 [Jatropha curcas] |
| 15 |
Hb_000325_070 |
0.1311907026 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_035677_010 |
0.1328257766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001677_120 |
0.1335444318 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 18 |
Hb_000024_020 |
0.1352306836 |
- |
- |
PREDICTED: uncharacterized protein LOC105636896 [Jatropha curcas] |
| 19 |
Hb_000247_150 |
0.1364776178 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 20 |
Hb_134022_010 |
0.1369207227 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |