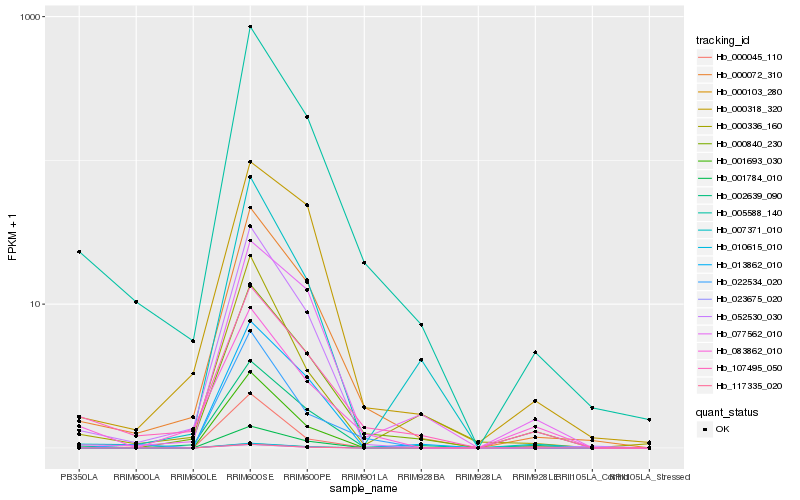
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000072_310 |
0.071890937 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 3 |
Hb_107495_050 |
0.1102131826 |
transcription factor |
TF Family: MIKC |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_052530_030 |
0.1206046304 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 5 |
Hb_002639_090 |
0.123370866 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 6 |
Hb_000840_230 |
0.1430549159 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 7 |
Hb_022534_020 |
0.158539766 |
- |
- |
PREDICTED: uncharacterized protein LOC103422158 [Malus domestica] |
| 8 |
Hb_000336_160 |
0.1632977971 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_000318_320 |
0.1679845453 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 10 |
Hb_007371_010 |
0.1705676271 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT-like [Populus euphratica] |
| 11 |
Hb_083862_010 |
0.1710835964 |
- |
- |
hypothetical protein JCGZ_17956 [Jatropha curcas] |
| 12 |
Hb_013862_010 |
0.1725001621 |
- |
- |
PREDICTED: uncharacterized protein LOC105650642 [Jatropha curcas] |
| 13 |
Hb_001693_030 |
0.1730153343 |
- |
- |
PREDICTED: reticuline oxidase-like [Jatropha curcas] |
| 14 |
Hb_077562_010 |
0.173571491 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 15 |
Hb_023675_020 |
0.173737043 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 16 |
Hb_000103_280 |
0.1741262458 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 17 |
Hb_010615_010 |
0.1746531026 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 18 |
Hb_000045_110 |
0.1750334978 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 19 |
Hb_001784_010 |
0.1753618872 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 20 |
Hb_117335_020 |
0.1755782452 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |