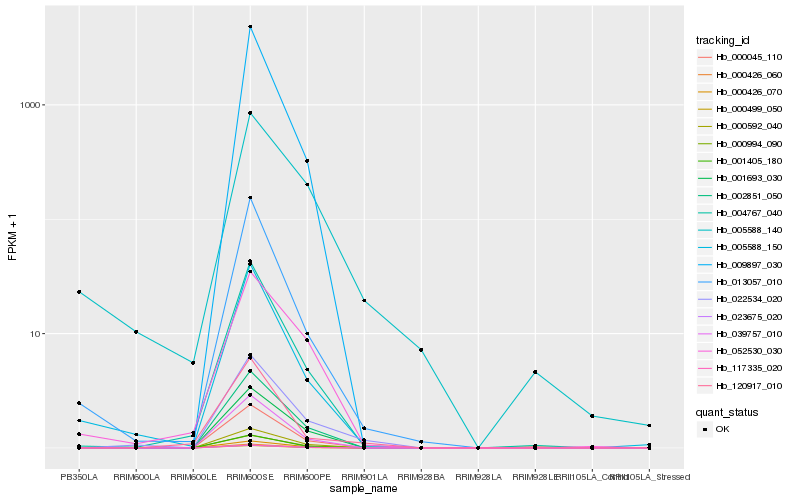
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022534_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103422158 [Malus domestica] |
| 2 |
Hb_000045_110 |
0.127330845 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 3 |
Hb_000426_060 |
0.1330222761 |
- |
- |
hypothetical protein CISIN_1g048192mg, partial [Citrus sinensis] |
| 4 |
Hb_002851_050 |
0.1331053435 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 5 |
Hb_000994_090 |
0.1335656124 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 6 |
Hb_000426_070 |
0.134077708 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 7 |
Hb_000592_040 |
0.1341881733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001405_180 |
0.1359228839 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000499_050 |
0.1359354437 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_039757_010 |
0.1367898697 |
- |
- |
- |
| 11 |
Hb_001693_030 |
0.1434640015 |
- |
- |
PREDICTED: reticuline oxidase-like [Jatropha curcas] |
| 12 |
Hb_004767_040 |
0.1455666743 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_117335_020 |
0.1456339735 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 14 |
Hb_023675_020 |
0.1537012323 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 15 |
Hb_009897_030 |
0.1553925064 |
- |
- |
- |
| 16 |
Hb_013057_010 |
0.1559232787 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 43-like [Jatropha curcas] |
| 17 |
Hb_005588_150 |
0.1565214825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005588_140 |
0.158539766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_052530_030 |
0.163273031 |
- |
- |
PREDICTED: cytochrome P450 78A9 [Jatropha curcas] |
| 20 |
Hb_120917_010 |
0.165114428 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |