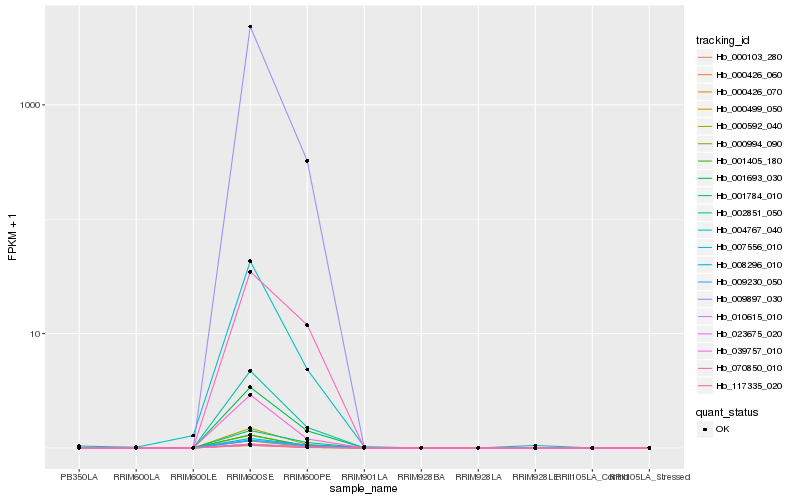
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000499_050 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_000592_040 |
0.0103511054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000426_070 |
0.0112069477 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Jatropha curcas] |
| 4 |
Hb_000994_090 |
0.0159034257 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 5 |
Hb_002851_050 |
0.0227930063 |
transcription factor |
TF Family: MYB-related |
myb family transcription factor family protein [Populus trichocarpa] |
| 6 |
Hb_000426_060 |
0.031422776 |
- |
- |
hypothetical protein CISIN_1g048192mg, partial [Citrus sinensis] |
| 7 |
Hb_117335_020 |
0.0315659079 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 8 |
Hb_001693_030 |
0.0406808581 |
- |
- |
PREDICTED: reticuline oxidase-like [Jatropha curcas] |
| 9 |
Hb_023675_020 |
0.0494993598 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 10 |
Hb_001405_180 |
0.0566851504 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_039757_010 |
0.06063753 |
- |
- |
- |
| 12 |
Hb_000103_280 |
0.0740529602 |
transcription factor |
TF Family: HSF |
Heat shock factor protein HSF30, putative [Ricinus communis] |
| 13 |
Hb_010615_010 |
0.0793505939 |
- |
- |
PREDICTED: reticuline oxidase-like protein [Jatropha curcas] |
| 14 |
Hb_001784_010 |
0.0848335658 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 15 |
Hb_008296_010 |
0.0863111755 |
- |
- |
- |
| 16 |
Hb_009230_050 |
0.0916826659 |
- |
- |
PREDICTED: transcription factor bHLH113-like [Jatropha curcas] |
| 17 |
Hb_004767_040 |
0.0997444532 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 18 |
Hb_009897_030 |
0.1090677268 |
- |
- |
- |
| 19 |
Hb_070850_010 |
0.1134017085 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B-like [Jatropha curcas] |
| 20 |
Hb_007556_010 |
0.1144208132 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X2 [Populus euphratica] |