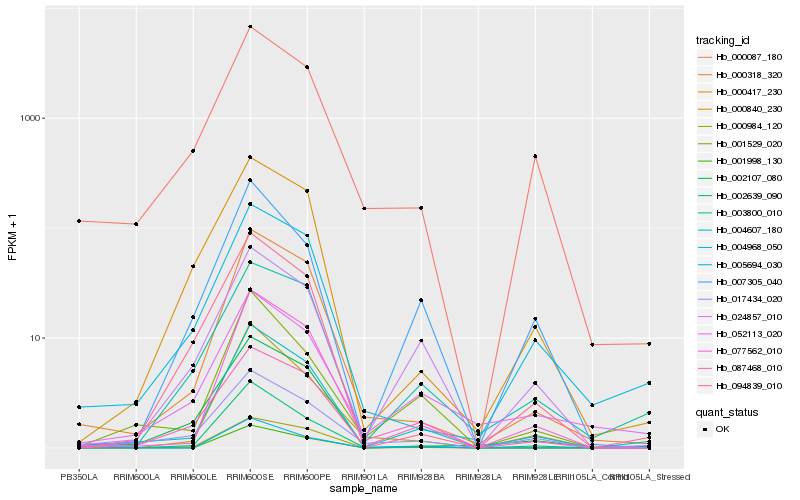
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004607_180 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_077562_010 |
0.1012604579 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 3 |
Hb_000318_320 |
0.1246878599 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 4 |
Hb_002639_090 |
0.1327546657 |
- |
- |
hypothetical protein JCGZ_08134 [Jatropha curcas] |
| 5 |
Hb_087468_010 |
0.1333993581 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 6 |
Hb_017434_020 |
0.1344271563 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 7 |
Hb_001529_020 |
0.1407843226 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003800_010 |
0.1436050788 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 9 |
Hb_000984_120 |
0.1463837559 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000840_230 |
0.1491854139 |
- |
- |
RING-H2 finger protein ATL1L, putative [Ricinus communis] |
| 11 |
Hb_024857_010 |
0.1506091734 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 12 |
Hb_001998_130 |
0.1510709038 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 35 family protein [Populus trichocarpa] |
| 13 |
Hb_002107_080 |
0.1518669307 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 14 |
Hb_000417_230 |
0.1547805087 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004968_050 |
0.1550467043 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 16 |
Hb_000087_180 |
0.1566641969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_052113_020 |
0.1584284241 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 18 |
Hb_007305_040 |
0.1595763516 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 19 |
Hb_094839_010 |
0.1597761158 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005694_030 |
0.1616642344 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PUB22-like [Jatropha curcas] |