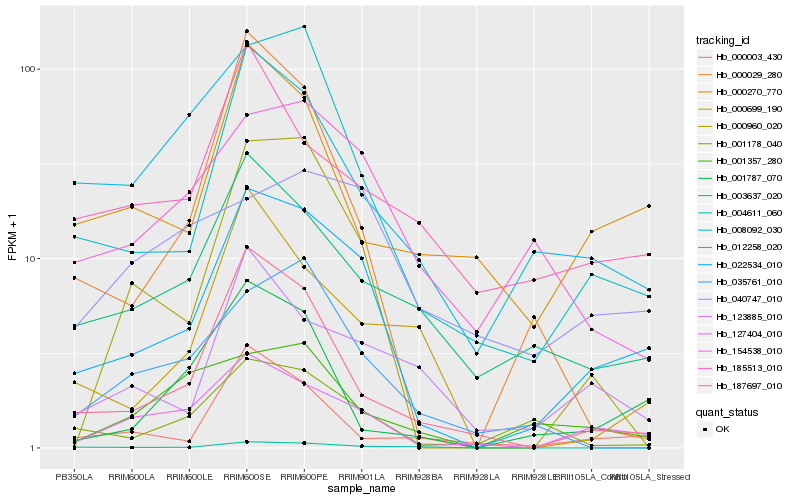
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022534_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10000066mg [Citrus clementina] |
| 2 |
Hb_127404_010 |
0.1775238109 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X1 [Gossypium raimondii] |
| 3 |
Hb_008092_030 |
0.1916828748 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 4 |
Hb_187697_010 |
0.2205818361 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 5 |
Hb_003637_020 |
0.2271484606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000960_020 |
0.2298884396 |
- |
- |
- |
| 7 |
Hb_001178_040 |
0.2342220643 |
desease resistance |
Gene Name: ABC_trans_N |
PREDICTED: ABC transporter G family member 34-like [Jatropha curcas] |
| 8 |
Hb_154538_010 |
0.2366819627 |
- |
- |
- |
| 9 |
Hb_123885_010 |
0.2446812141 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_000270_770 |
0.2463126114 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 11 |
Hb_000003_430 |
0.2543193032 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 12 |
Hb_001787_070 |
0.2546533234 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Camelina sativa] |
| 13 |
Hb_012258_020 |
0.2578304136 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_004611_060 |
0.2583643789 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 15 |
Hb_185513_010 |
0.2599053157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000029_280 |
0.2610900204 |
- |
- |
PREDICTED: uncharacterized protein LOC105631804 [Jatropha curcas] |
| 17 |
Hb_000699_190 |
0.2632285646 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_035761_010 |
0.2654447207 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 19 |
Hb_001357_280 |
0.2685954364 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 20 |
Hb_040747_010 |
0.2693818112 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |