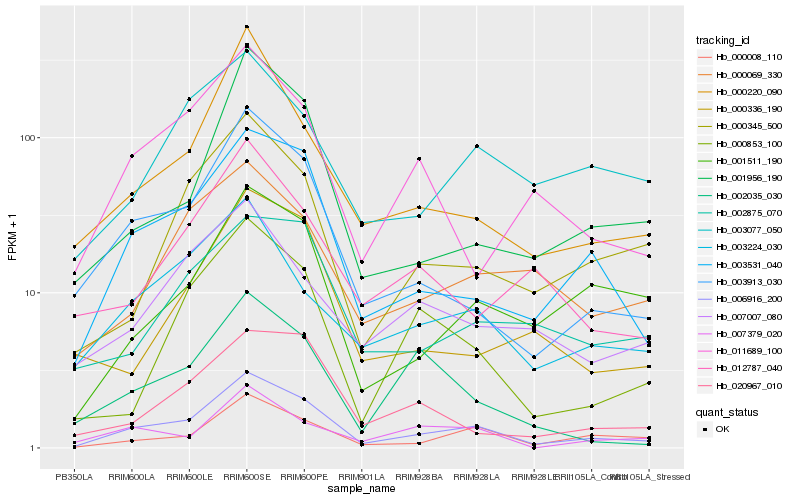
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_200 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 2 |
Hb_003531_040 |
0.1383832238 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 3 |
Hb_003913_030 |
0.1520599293 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 4 |
Hb_000008_110 |
0.161118187 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 5 |
Hb_000345_500 |
0.1790536144 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 6 |
Hb_000220_090 |
0.1798847874 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 7 |
Hb_001511_190 |
0.1819282437 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 8 |
Hb_001956_190 |
0.1824164647 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 9 |
Hb_003224_030 |
0.1846398443 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 10 |
Hb_000853_100 |
0.1897629193 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 11 |
Hb_020967_010 |
0.192664026 |
- |
- |
- |
| 12 |
Hb_011689_100 |
0.1959220074 |
- |
- |
PREDICTED: uncharacterized protein LOC105631864 [Jatropha curcas] |
| 13 |
Hb_002875_070 |
0.1966215661 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 14 |
Hb_000069_330 |
0.1967679077 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_003077_050 |
0.1990681799 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 16 |
Hb_000336_190 |
0.2000550262 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 17 |
Hb_007379_020 |
0.2047323809 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 18 |
Hb_007007_080 |
0.2058382109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_012787_040 |
0.2090601629 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002035_030 |
0.2105056392 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |