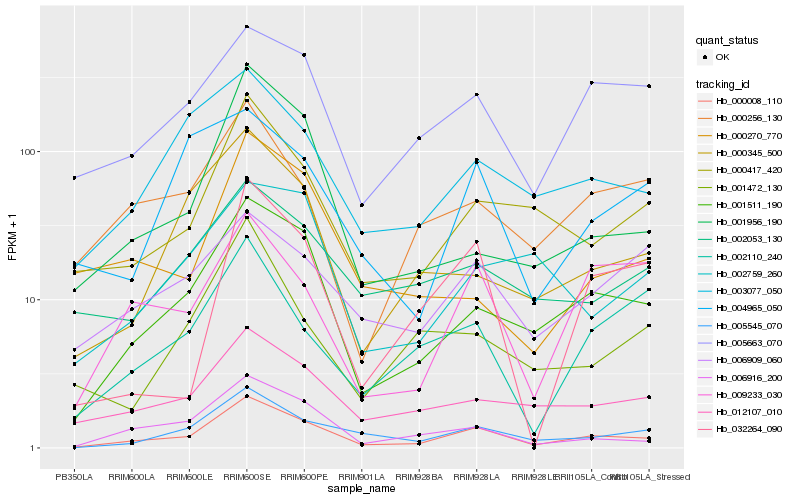
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_110 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 2 |
Hb_001511_190 |
0.128949867 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 3 |
Hb_005545_070 |
0.1577140472 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 4 |
Hb_006916_200 |
0.161118187 |
- |
- |
hypothetical protein POPTR_0017s13190g, partial [Populus trichocarpa] |
| 5 |
Hb_000417_420 |
0.1793195401 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 6 |
Hb_005663_070 |
0.1822711225 |
- |
- |
dehydrin protein [Manihot esculenta] |
| 7 |
Hb_012107_010 |
0.1829899068 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 8 |
Hb_003077_050 |
0.1848458887 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 9 |
Hb_000345_500 |
0.1996818696 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 10 |
Hb_009233_030 |
0.2021916335 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_001956_190 |
0.2046592374 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 12 |
Hb_000256_130 |
0.2054277662 |
- |
- |
hypothetical protein 21 [Hevea brasiliensis] |
| 13 |
Hb_002053_130 |
0.2064296522 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 14 |
Hb_002110_240 |
0.2128423427 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 15 |
Hb_032264_090 |
0.2144833774 |
- |
- |
hypothetical protein JCGZ_02512 [Jatropha curcas] |
| 16 |
Hb_006909_060 |
0.2173471886 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 17 |
Hb_001472_130 |
0.2204356017 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 18 |
Hb_002759_260 |
0.2220012606 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 19 |
Hb_000270_770 |
0.2229274019 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 20 |
Hb_004965_050 |
0.2233684303 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |