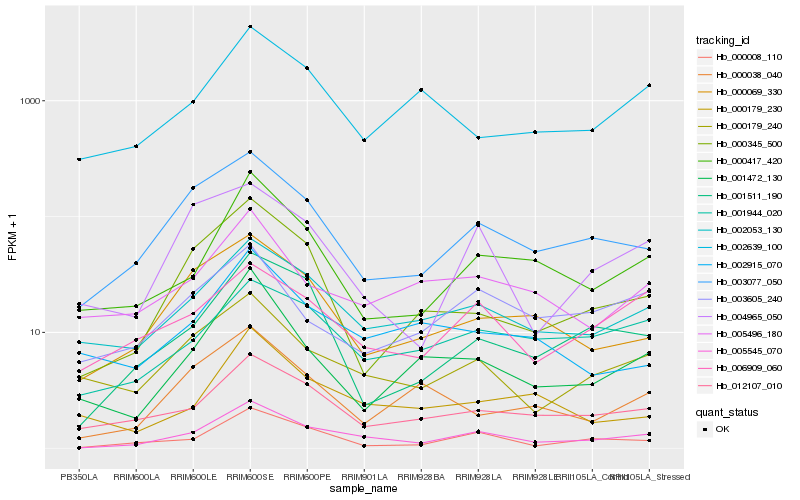
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005545_070 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_000008_110 |
0.1577140472 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Gossypium raimondii] |
| 3 |
Hb_002053_130 |
0.1629000923 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 4 |
Hb_003077_050 |
0.1683826624 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 5 |
Hb_000417_420 |
0.1745411749 |
- |
- |
PREDICTED: U-box domain-containing protein 17 [Jatropha curcas] |
| 6 |
Hb_012107_010 |
0.1762708782 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 7 |
Hb_000179_240 |
0.1841298716 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001511_190 |
0.1847806131 |
- |
- |
PREDICTED: probable protein phosphatase 2C 25 [Jatropha curcas] |
| 9 |
Hb_001944_020 |
0.187520244 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 10 |
Hb_000069_330 |
0.1910952993 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_006909_060 |
0.1924251296 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3-like [Solanum lycopersicum] |
| 12 |
Hb_001472_130 |
0.1956049718 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 13 |
Hb_000345_500 |
0.1963410861 |
- |
- |
PREDICTED: peroxisomal adenine nucleotide carrier 1 [Jatropha curcas] |
| 14 |
Hb_004965_050 |
0.2021415257 |
- |
- |
PREDICTED: uncharacterized protein LOC105645055 [Jatropha curcas] |
| 15 |
Hb_000179_230 |
0.2068944169 |
- |
- |
PREDICTED: myosin-17 [Jatropha curcas] |
| 16 |
Hb_005496_180 |
0.2073430413 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 17 |
Hb_003605_240 |
0.2084717184 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 18 |
Hb_002639_100 |
0.2097959589 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 19 |
Hb_000038_040 |
0.2151319653 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 20 |
Hb_002915_070 |
0.2152377695 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |