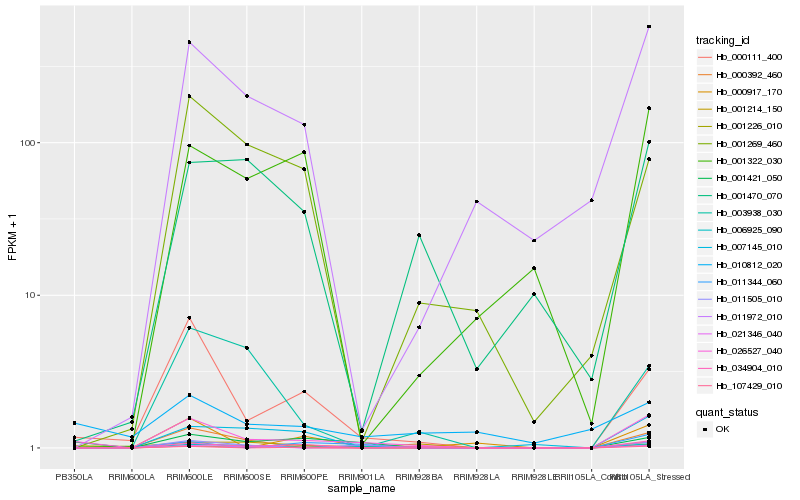
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026527_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001421_050 |
0.259454149 |
- |
- |
- |
| 3 |
Hb_000111_400 |
0.2703747157 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 4 |
Hb_006925_090 |
0.2950866228 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 5 |
Hb_034904_010 |
0.2988287313 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 6 |
Hb_011972_010 |
0.3175275846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011505_010 |
0.3270006842 |
- |
- |
- |
| 8 |
Hb_001226_010 |
0.3301415446 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001214_150 |
0.3326988575 |
- |
- |
PREDICTED: uncharacterized protein LOC102624288 [Citrus sinensis] |
| 10 |
Hb_007145_010 |
0.3398422689 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 11 |
Hb_001322_030 |
0.3448344118 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_010812_020 |
0.3466145799 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 13 |
Hb_000392_460 |
0.3502290809 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 14 |
Hb_001470_070 |
0.3517541609 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_011344_060 |
0.3521905262 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000917_170 |
0.3538328937 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 17 |
Hb_107429_010 |
0.3563921221 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 18 |
Hb_021346_040 |
0.3581302019 |
- |
- |
hypothetical protein JCGZ_26751 [Jatropha curcas] |
| 19 |
Hb_003938_030 |
0.3606848807 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |
| 20 |
Hb_001269_460 |
0.3630577438 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |