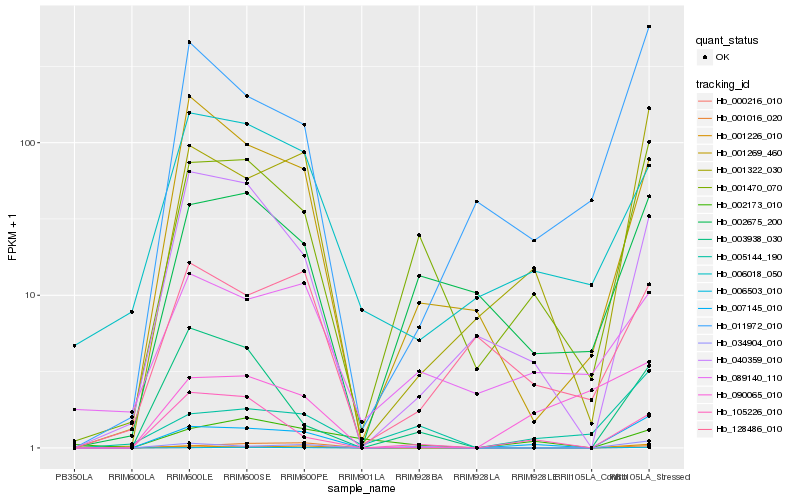
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007145_010 |
0.0 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 2 |
Hb_001322_030 |
0.1224948563 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 3 |
Hb_001470_070 |
0.1973874982 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 4 |
Hb_001226_010 |
0.2182222999 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_011972_010 |
0.2201849936 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_034904_010 |
0.2462922158 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 7 |
Hb_040359_010 |
0.2507976833 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 8 |
Hb_105226_010 |
0.2613689398 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 9 |
Hb_005144_190 |
0.2710588012 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 10 |
Hb_002675_200 |
0.272872753 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 11 |
Hb_001269_460 |
0.2800551503 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 12 |
Hb_128486_010 |
0.2846934107 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 13 |
Hb_006018_050 |
0.2869859243 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 14 |
Hb_006503_010 |
0.2897739657 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_090065_010 |
0.2935589999 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_089140_110 |
0.2995648054 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002173_010 |
0.3093936318 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 18 |
Hb_001016_020 |
0.3104737323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000216_010 |
0.3113431623 |
- |
- |
carbon monoxide dehydrogenase [Streptomyces sp. CNQ-509] |
| 20 |
Hb_003938_030 |
0.3133331216 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 2 [Hevea brasiliensis] |