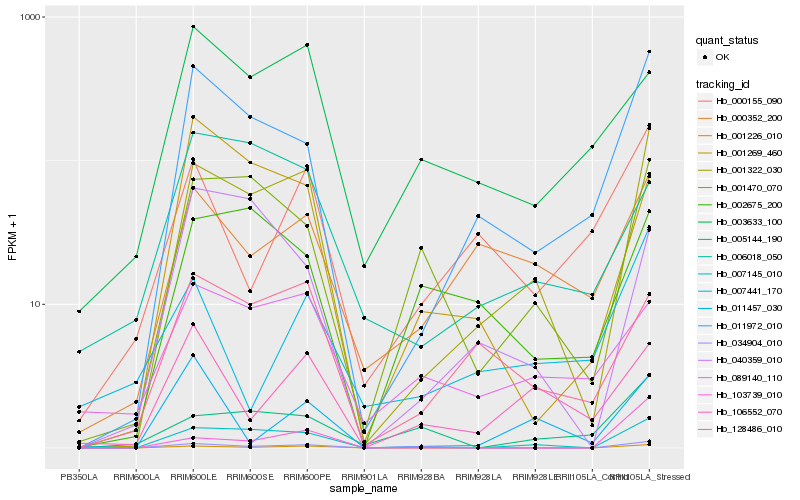
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001322_030 |
0.0 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 2 |
Hb_007145_010 |
0.1224948563 |
- |
- |
ammonium transporter 3.1-like protein [Camellia sinensis] |
| 3 |
Hb_001226_010 |
0.1844732968 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011972_010 |
0.1914873501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001470_070 |
0.2325810372 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_034904_010 |
0.2391849836 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 7 |
Hb_128486_010 |
0.240560702 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 8 |
Hb_000352_200 |
0.259507472 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002675_200 |
0.2784417895 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA1, putative [Ricinus communis] |
| 10 |
Hb_089140_110 |
0.2808719264 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005144_190 |
0.2817018463 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 12 |
Hb_106552_070 |
0.28508581 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_103739_010 |
0.2859690159 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 14 |
Hb_040359_010 |
0.2884377679 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 15 |
Hb_000155_090 |
0.289540419 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 16 |
Hb_001269_460 |
0.2944680635 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 17 |
Hb_003633_100 |
0.2960973392 |
- |
- |
hypoia-responsive family protein 1 [Hevea brasiliensis] |
| 18 |
Hb_006018_050 |
0.3038116543 |
- |
- |
PREDICTED: probable protein phosphatase 2C 63 [Jatropha curcas] |
| 19 |
Hb_011457_030 |
0.3103107603 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 20 |
Hb_007441_170 |
0.313943022 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |