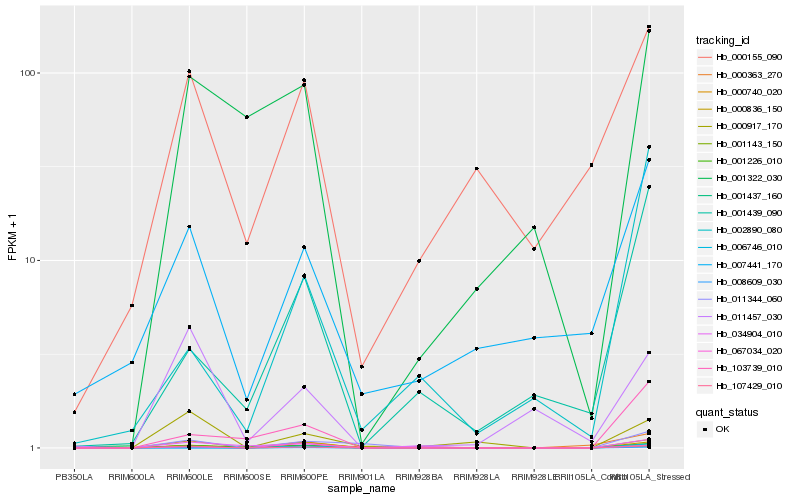
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_160 |
0.0 |
- |
- |
PREDICTED: high-affinity nitrate transporter 2.1-like [Jatropha curcas] |
| 2 |
Hb_000740_020 |
0.1306631021 |
transcription factor |
TF Family: MYB-related |
asymmetric leaves1 and rough sheath, putative [Ricinus communis] |
| 3 |
Hb_107429_010 |
0.1477810223 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 4 |
Hb_000836_150 |
0.2023204634 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 5 |
Hb_001226_010 |
0.2110097774 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_103739_010 |
0.2427725513 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 7 |
Hb_011344_060 |
0.2507683522 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X2 [Jatropha curcas] |
| 8 |
Hb_067034_020 |
0.2810048773 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |
| 9 |
Hb_001439_090 |
0.2900645173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001143_150 |
0.2973246356 |
- |
- |
PREDICTED: mitochondrial protein import protein MAS5-like [Populus euphratica] |
| 11 |
Hb_034904_010 |
0.3107928465 |
- |
- |
PREDICTED: uncharacterized protein LOC105775231 [Gossypium raimondii] |
| 12 |
Hb_000917_170 |
0.3116523777 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 13 |
Hb_002890_080 |
0.3162126778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001322_030 |
0.3208206292 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 15 |
Hb_007441_170 |
0.3284902603 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 16 |
Hb_000363_270 |
0.3386175595 |
- |
- |
hypothetical protein PRUPE_ppa024846mg [Prunus persica] |
| 17 |
Hb_000155_090 |
0.3436314211 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 18 |
Hb_011457_030 |
0.3443184872 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 19 |
Hb_008609_030 |
0.350192096 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_006746_010 |
0.3503692512 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |