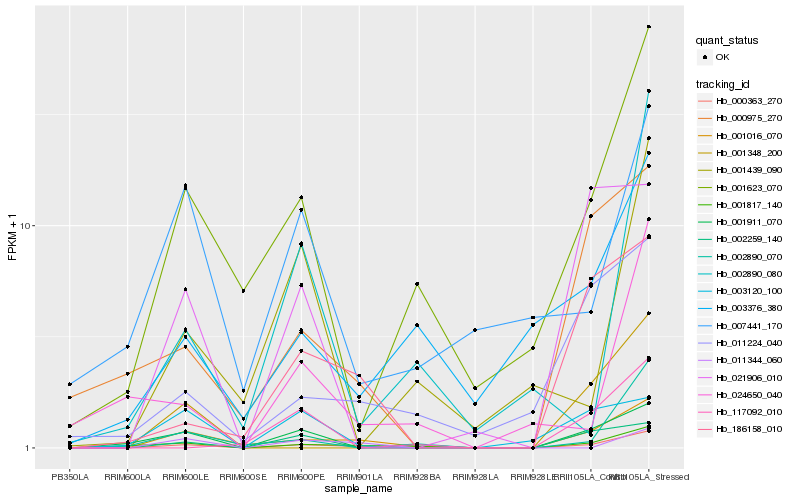
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_270 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa024846mg [Prunus persica] |
| 2 |
Hb_001016_070 |
0.1747024798 |
- |
- |
PREDICTED: putative MO25-like protein At5g47540 [Jatropha curcas] |
| 3 |
Hb_001817_140 |
0.2167572333 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_186158_010 |
0.2634110689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001623_070 |
0.2638303586 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 6 |
Hb_002259_140 |
0.2716529002 |
- |
- |
- |
| 7 |
Hb_001911_070 |
0.2722897021 |
- |
- |
- |
| 8 |
Hb_000975_270 |
0.2786412731 |
- |
- |
PREDICTED: histone H2B.11-like [Phoenix dactylifera] |
| 9 |
Hb_002890_070 |
0.2828640643 |
- |
- |
ATP-dependent RNA helicase family protein [Populus trichocarpa] |
| 10 |
Hb_011344_060 |
0.3021028685 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001348_200 |
0.3027431199 |
- |
- |
- |
| 12 |
Hb_011224_040 |
0.3035853392 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 13 |
Hb_021906_010 |
0.3164959122 |
- |
- |
PREDICTED: cysteine proteinase inhibitor-like [Populus euphratica] |
| 14 |
Hb_024650_040 |
0.3233345217 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 15 |
Hb_003376_380 |
0.3250024582 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 16 |
Hb_001439_090 |
0.3266371931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002890_080 |
0.3286067035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_117092_010 |
0.3286126583 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 19 |
Hb_003120_100 |
0.3290184923 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 20 |
Hb_007441_170 |
0.330355966 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |