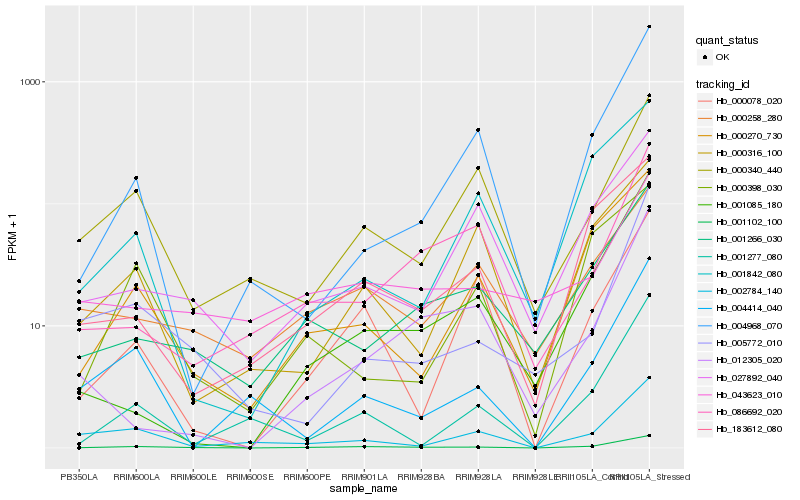
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105140429 isoform X2 [Populus euphratica] |
| 2 |
Hb_000270_730 |
0.1956599487 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 3 |
Hb_001266_030 |
0.2041143168 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_001277_080 |
0.2064923643 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 5 |
Hb_005772_010 |
0.207515968 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 6 |
Hb_183612_080 |
0.2080954392 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 7 |
Hb_002784_140 |
0.2153126696 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 8 |
Hb_004414_040 |
0.2153491438 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 9 |
Hb_001085_180 |
0.2163832323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000340_440 |
0.220607349 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 11 |
Hb_000078_020 |
0.2219750586 |
- |
- |
- |
| 12 |
Hb_027892_040 |
0.2249777286 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 13 |
Hb_004968_070 |
0.2253877514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000258_280 |
0.2300486419 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 15 |
Hb_086692_020 |
0.2392896959 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000316_100 |
0.2418567159 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_012305_020 |
0.2421357372 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 18 |
Hb_001842_080 |
0.2434725391 |
- |
- |
- |
| 19 |
Hb_000398_030 |
0.244776365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_043623_010 |
0.244989939 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |