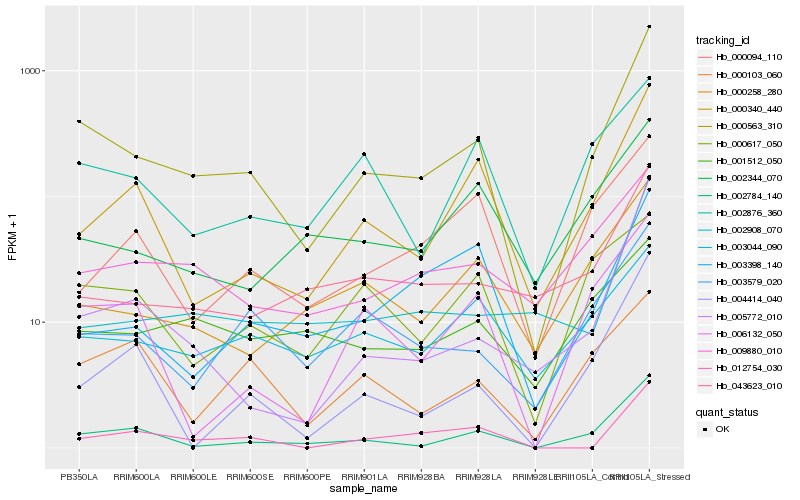
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_310 |
0.0 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 2 |
Hb_002784_140 |
0.1675656736 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_000340_440 |
0.1935069086 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 4 |
Hb_005772_010 |
0.1969676798 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 5 |
Hb_003579_020 |
0.2014607672 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 6 |
Hb_000258_280 |
0.2083274959 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 7 |
Hb_009880_010 |
0.2085643654 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 8 |
Hb_006132_050 |
0.2108315004 |
- |
- |
hypothetical protein EUTSA_v10028860mg [Eutrema salsugineum] |
| 9 |
Hb_002908_070 |
0.2118242667 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 10 |
Hb_043623_010 |
0.2128542277 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 11 |
Hb_004414_040 |
0.2171816554 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 12 |
Hb_002876_360 |
0.2190418812 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 13 |
Hb_012754_030 |
0.2253666309 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002344_070 |
0.2272695274 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 15 |
Hb_003044_090 |
0.2295240936 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 16 |
Hb_003398_140 |
0.2343515521 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000094_110 |
0.2361285816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000103_060 |
0.2363322145 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 19 |
Hb_001512_050 |
0.237090258 |
- |
- |
copine, putative [Ricinus communis] |
| 20 |
Hb_000617_050 |
0.2382131746 |
- |
- |
PREDICTED: uncharacterized protein LOC105647466 [Jatropha curcas] |