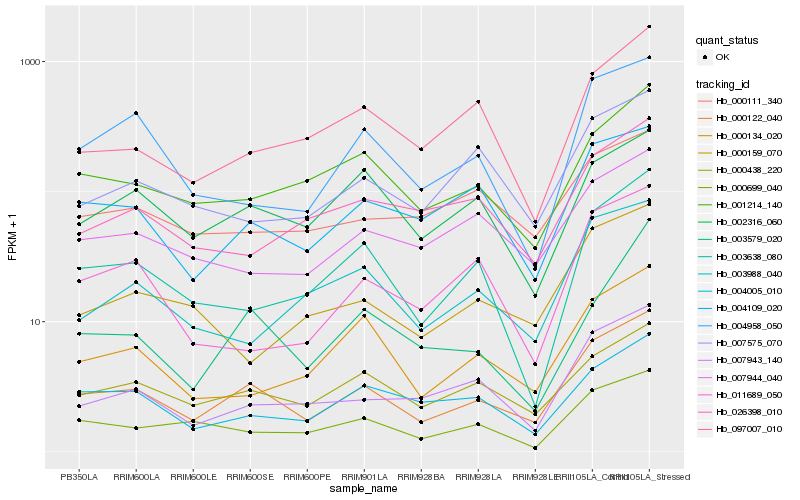
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649224 [Jatropha curcas] |
| 2 |
Hb_007943_140 |
0.1320400152 |
- |
- |
PREDICTED: uncharacterized protein LOC103340977 [Prunus mume] |
| 3 |
Hb_000438_220 |
0.1338429785 |
- |
- |
PREDICTED: uncharacterized protein LOC105649213 [Jatropha curcas] |
| 4 |
Hb_004109_020 |
0.1360661739 |
- |
- |
60S ribosomal protein L18aA [Hevea brasiliensis] |
| 5 |
Hb_001214_140 |
0.1390874628 |
- |
- |
60S ribosomal protein L27a, putative [Ricinus communis] |
| 6 |
Hb_003638_080 |
0.1423409347 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 7 |
Hb_000699_040 |
0.1435911332 |
- |
- |
PREDICTED: uncharacterized protein LOC105647732 [Jatropha curcas] |
| 8 |
Hb_000134_020 |
0.1448794078 |
- |
- |
thioredoxin-like family protein [Hevea brasiliensis] |
| 9 |
Hb_004958_050 |
0.1502514382 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 10 |
Hb_004005_010 |
0.150596882 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 11 |
Hb_097007_010 |
0.152656257 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 12 |
Hb_003988_040 |
0.1538189291 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 13 |
Hb_007944_040 |
0.1546985828 |
- |
- |
60S ribosomal protein L18aB [Hevea brasiliensis] |
| 14 |
Hb_007575_070 |
0.1550060717 |
- |
- |
Histone H3.2 [Glycine soja] |
| 15 |
Hb_000159_070 |
0.1588527577 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 16 |
Hb_003579_020 |
0.1610154629 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 17 |
Hb_002316_060 |
0.1624216604 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 18 |
Hb_011689_050 |
0.1624640741 |
- |
- |
hypothetical protein POPTR_0008s20290g [Populus trichocarpa] |
| 19 |
Hb_026398_010 |
0.1629632757 |
- |
- |
hypothetical protein B456_010G132100 [Gossypium raimondii] |
| 20 |
Hb_000111_340 |
0.1662673237 |
- |
- |
peptidyl-prolyl cis-trans isomerase h, ppih, putative [Ricinus communis] |