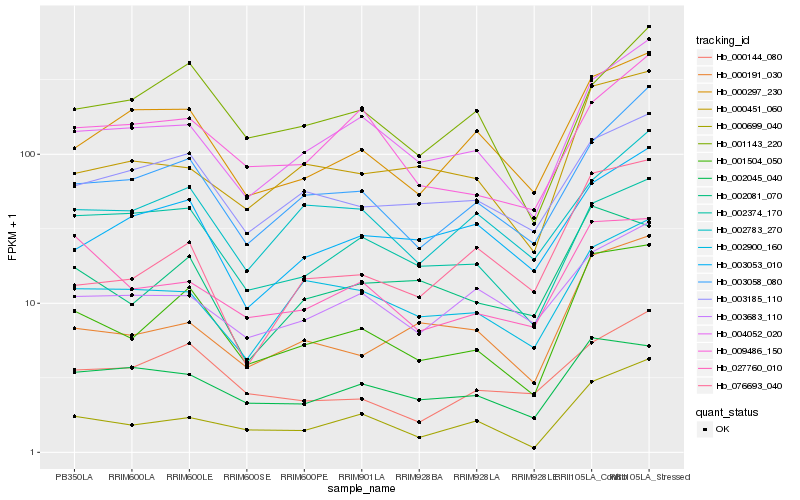
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s14840g [Populus trichocarpa] |
| 2 |
Hb_000699_040 |
0.1200591708 |
- |
- |
PREDICTED: uncharacterized protein LOC105647732 [Jatropha curcas] |
| 3 |
Hb_004052_020 |
0.1217412049 |
- |
- |
Mitochondrial import receptor subunit TOM5 [Theobroma cacao] |
| 4 |
Hb_003058_080 |
0.1285907359 |
- |
- |
Chaperonin 10 isoform 1 [Theobroma cacao] |
| 5 |
Hb_000451_060 |
0.1315913148 |
- |
- |
hypothetical protein CICLE_v10010074mg [Citrus clementina] |
| 6 |
Hb_000191_030 |
0.1325602392 |
- |
- |
hypothetical protein POPTR_0012s07830g [Populus trichocarpa] |
| 7 |
Hb_000297_230 |
0.1333157755 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 8 |
Hb_002081_070 |
0.1365709621 |
- |
- |
PREDICTED: pyridoxal kinase [Jatropha curcas] |
| 9 |
Hb_003185_110 |
0.1369935827 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 7-2 [Nelumbo nucifera] |
| 10 |
Hb_027760_010 |
0.1417897035 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 11 |
Hb_002900_160 |
0.146003428 |
- |
- |
PREDICTED: immediate early response 3-interacting protein 1-like [Jatropha curcas] |
| 12 |
Hb_000144_080 |
0.1465481915 |
- |
- |
PREDICTED: uncharacterized protein LOC104609774 [Nelumbo nucifera] |
| 13 |
Hb_002045_040 |
0.1506162384 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002374_170 |
0.1535036688 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |
| 15 |
Hb_003053_010 |
0.1539500717 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 16 |
Hb_009486_150 |
0.1543227074 |
- |
- |
60S ribosomal protein L37a-2 [Medicago truncatula] |
| 17 |
Hb_002783_270 |
0.1543611131 |
- |
- |
PREDICTED: AP-2 complex subunit sigma [Fragaria vesca subsp. vesca] |
| 18 |
Hb_076693_040 |
0.1550536867 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 19 |
Hb_001143_220 |
0.1559407589 |
- |
- |
20S proteasome beta subunit C2 [Hevea brasiliensis] |
| 20 |
Hb_003683_110 |
0.1561318244 |
- |
- |
JHL10I11.3 [Jatropha curcas] |