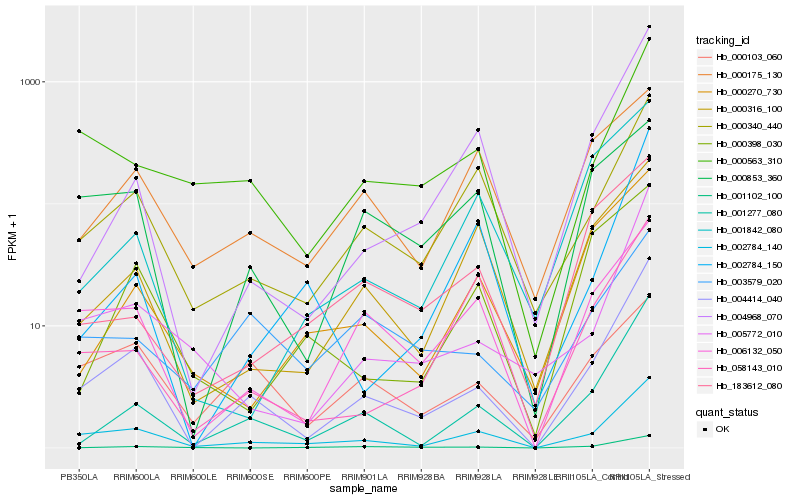
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004414_040 |
0.0 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_002784_140 |
0.1275276346 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 3 |
Hb_001277_080 |
0.1622583569 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 4 |
Hb_005772_010 |
0.1951196699 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 5 |
Hb_000340_440 |
0.1958400853 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 6 |
Hb_006132_050 |
0.2074996078 |
- |
- |
hypothetical protein EUTSA_v10028860mg [Eutrema salsugineum] |
| 7 |
Hb_004968_070 |
0.2127638953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001102_100 |
0.2153491438 |
- |
- |
PREDICTED: uncharacterized protein LOC105140429 isoform X2 [Populus euphratica] |
| 9 |
Hb_000563_310 |
0.2171816554 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 10 |
Hb_000316_100 |
0.236814567 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000270_730 |
0.2419960316 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 12 |
Hb_000853_360 |
0.2495915982 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 13 |
Hb_002784_150 |
0.252282137 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 14 |
Hb_058143_010 |
0.2526538345 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 15 |
Hb_003579_020 |
0.2552063284 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 16 |
Hb_183612_080 |
0.2558448247 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 17 |
Hb_000103_060 |
0.2564275272 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 18 |
Hb_001842_080 |
0.2582857274 |
- |
- |
- |
| 19 |
Hb_000175_130 |
0.2601458123 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 20 |
Hb_000398_030 |
0.262346151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |