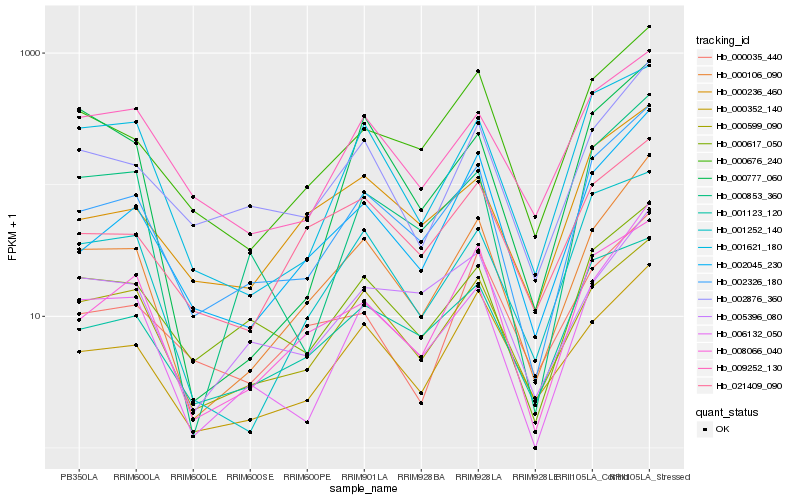
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000106_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 2 |
Hb_002326_180 |
0.0991950323 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 3 |
Hb_000352_140 |
0.1196441994 |
- |
- |
- |
| 4 |
Hb_002045_230 |
0.1222732301 |
- |
- |
Ammonium transporter 2 [Theobroma cacao] |
| 5 |
Hb_006132_050 |
0.1232357016 |
- |
- |
hypothetical protein EUTSA_v10028860mg [Eutrema salsugineum] |
| 6 |
Hb_002876_360 |
0.123667091 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 7 |
Hb_000599_090 |
0.125579887 |
- |
- |
hypothetical protein POPTR_0002s039001g, partial [Populus trichocarpa] |
| 8 |
Hb_000676_240 |
0.1292116472 |
- |
- |
thioredoxin H-type 2 [Hevea brasiliensis] |
| 9 |
Hb_008066_040 |
0.148651568 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL62 [Jatropha curcas] |
| 10 |
Hb_000617_050 |
0.1487553587 |
- |
- |
PREDICTED: uncharacterized protein LOC105647466 [Jatropha curcas] |
| 11 |
Hb_021409_090 |
0.1489659908 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0003s14450g [Populus trichocarpa] |
| 12 |
Hb_005396_080 |
0.1499917639 |
- |
- |
PREDICTED: nudix hydrolase 1 [Jatropha curcas] |
| 13 |
Hb_001252_140 |
0.1503900954 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_000853_360 |
0.150427344 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 15 |
Hb_001621_180 |
0.1534034052 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-1 [Jatropha curcas] |
| 16 |
Hb_000236_460 |
0.1535078083 |
- |
- |
PREDICTED: uncharacterized protein LOC105633119 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000035_440 |
0.1557235369 |
- |
- |
- |
| 18 |
Hb_001123_120 |
0.1593913573 |
- |
- |
Uncharacterized protein TCM_042776 [Theobroma cacao] |
| 19 |
Hb_009252_130 |
0.1594094117 |
- |
- |
Deletion of SUV3 suppressor 1(I) isoform 1 [Theobroma cacao] |
| 20 |
Hb_000777_060 |
0.1596349094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |