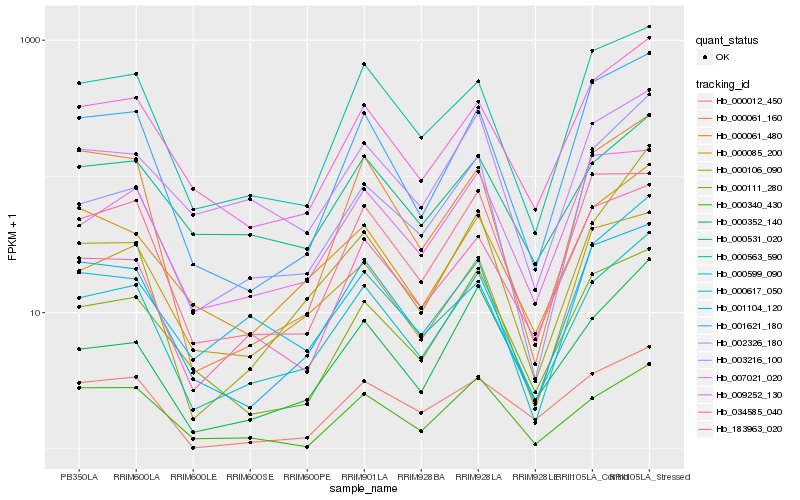
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s039001g, partial [Populus trichocarpa] |
| 2 |
Hb_009252_130 |
0.094410441 |
- |
- |
Deletion of SUV3 suppressor 1(I) isoform 1 [Theobroma cacao] |
| 3 |
Hb_000563_590 |
0.0990884203 |
- |
- |
PREDICTED: 60S ribosomal protein L22-2-like [Jatropha curcas] |
| 4 |
Hb_001621_180 |
0.1043841832 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-1 [Jatropha curcas] |
| 5 |
Hb_000061_480 |
0.1100996103 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 6 |
Hb_000111_280 |
0.1115557653 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 7 |
Hb_000531_020 |
0.1138256152 |
- |
- |
PREDICTED: uncharacterized protein At4g28440-like [Populus euphratica] |
| 8 |
Hb_000352_140 |
0.1175906174 |
- |
- |
- |
| 9 |
Hb_000085_200 |
0.1178877285 |
- |
- |
ATP synthase subunit H protein [Hevea brasiliensis] |
| 10 |
Hb_034585_040 |
0.1191404224 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 11 |
Hb_003216_100 |
0.1197212792 |
- |
- |
PREDICTED: uncharacterized protein LOC105641496 [Jatropha curcas] |
| 12 |
Hb_001104_120 |
0.1211386963 |
- |
- |
- |
| 13 |
Hb_000617_050 |
0.1218374911 |
- |
- |
PREDICTED: uncharacterized protein LOC105647466 [Jatropha curcas] |
| 14 |
Hb_007021_020 |
0.1229783417 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000061_160 |
0.1235622893 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_183963_020 |
0.1236424687 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 17 |
Hb_000012_450 |
0.1241980688 |
- |
- |
PREDICTED: uncharacterized protein LOC105638155 [Jatropha curcas] |
| 18 |
Hb_002326_180 |
0.1243584975 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 19 |
Hb_000340_430 |
0.1248509343 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000106_090 |
0.125579887 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |