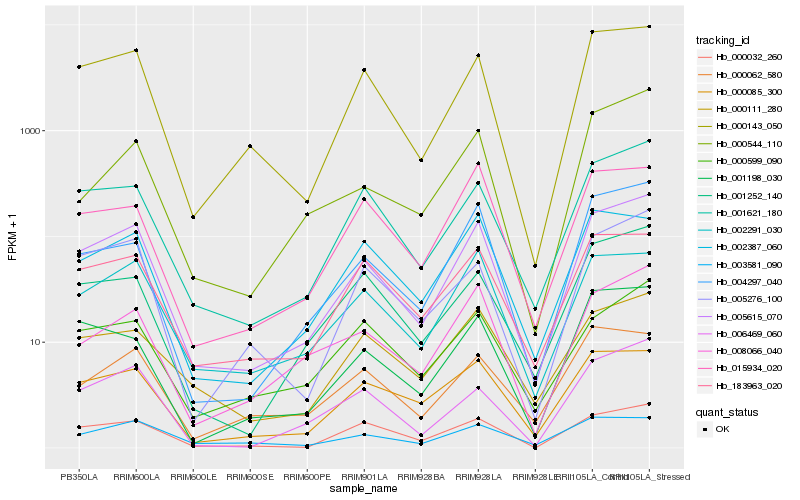
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005615_070 |
0.0 |
- |
- |
Rab7 [Hevea brasiliensis] |
| 2 |
Hb_001621_180 |
0.0960089915 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-1 [Jatropha curcas] |
| 3 |
Hb_000143_050 |
0.0985073759 |
- |
- |
PREDICTED: MFP1 attachment factor 1-like [Jatropha curcas] |
| 4 |
Hb_004297_040 |
0.1129956112 |
- |
- |
acyl CoA reductase [Hevea brasiliensis] |
| 5 |
Hb_000032_260 |
0.1152299472 |
- |
- |
PREDICTED: probable protein phosphatase 2C 51 [Jatropha curcas] |
| 6 |
Hb_002291_030 |
0.1183022718 |
- |
- |
PREDICTED: vesicle transport protein SFT2B [Jatropha curcas] |
| 7 |
Hb_000085_300 |
0.1184841064 |
- |
- |
- |
| 8 |
Hb_006469_060 |
0.1190033238 |
- |
- |
hypothetical protein POPTR_0005s16470g [Populus trichocarpa] |
| 9 |
Hb_005276_100 |
0.1212974636 |
- |
- |
PREDICTED: uncharacterized protein LOC105640215 [Jatropha curcas] |
| 10 |
Hb_008066_040 |
0.1229836872 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL62 [Jatropha curcas] |
| 11 |
Hb_001252_140 |
0.1235318731 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_183963_020 |
0.1258662105 |
- |
- |
PREDICTED: uncharacterized protein LOC105644615 [Jatropha curcas] |
| 13 |
Hb_001198_030 |
0.1271737826 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 14 |
Hb_000111_280 |
0.1287763222 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 15 |
Hb_002387_060 |
0.1351451666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000599_090 |
0.1359210011 |
- |
- |
hypothetical protein POPTR_0002s039001g, partial [Populus trichocarpa] |
| 17 |
Hb_015934_020 |
0.1375285533 |
- |
- |
PREDICTED: uncharacterized protein LOC105638669 [Jatropha curcas] |
| 18 |
Hb_000544_110 |
0.1381357727 |
- |
- |
PREDICTED: probable calcium-binding protein CML27 [Jatropha curcas] |
| 19 |
Hb_000062_580 |
0.1383856466 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_003581_090 |
0.1384674806 |
- |
- |
PREDICTED: galactose oxidase [Jatropha curcas] |