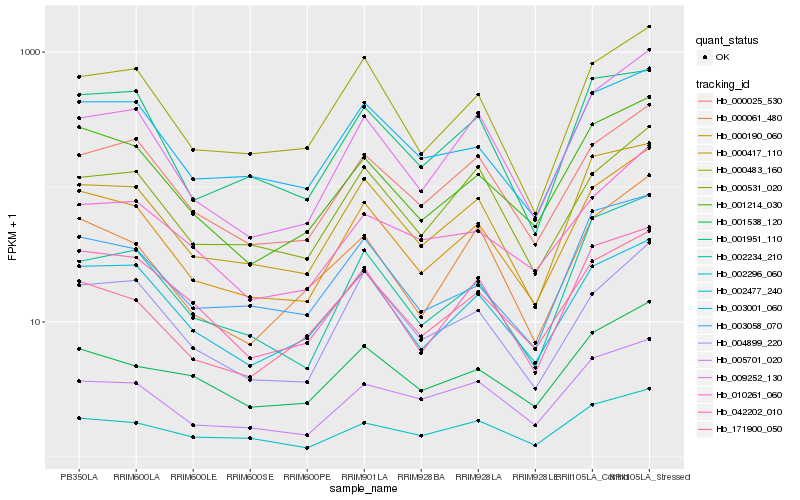
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000190_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644688 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001214_030 |
0.0636018049 |
- |
- |
hypothetical protein POPTR_0006s10280g [Populus trichocarpa] |
| 3 |
Hb_009252_130 |
0.0786793611 |
- |
- |
Deletion of SUV3 suppressor 1(I) isoform 1 [Theobroma cacao] |
| 4 |
Hb_000417_110 |
0.081108274 |
- |
- |
PREDICTED: uncharacterized protein LOC105649681 [Jatropha curcas] |
| 5 |
Hb_000483_160 |
0.0846273 |
- |
- |
PREDICTED: 60S ribosomal protein L34 [Jatropha curcas] |
| 6 |
Hb_000025_530 |
0.0879496061 |
- |
- |
PREDICTED: reactive oxygen species modulator 1 [Jatropha curcas] |
| 7 |
Hb_003058_070 |
0.0891549364 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 8 |
Hb_000061_480 |
0.0896133546 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 3 [Jatropha curcas] |
| 9 |
Hb_002234_210 |
0.0902709642 |
- |
- |
hypothetical protein POPTR_0012s13380g [Populus trichocarpa] |
| 10 |
Hb_171900_050 |
0.0905398611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003001_060 |
0.0908417093 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 12 |
Hb_004899_220 |
0.0914621924 |
- |
- |
PREDICTED: adrenodoxin-like protein 2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_002477_240 |
0.0926170628 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 14 |
Hb_042202_010 |
0.098500404 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 15 |
Hb_005701_020 |
0.1003835344 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 16 |
Hb_010261_060 |
0.100797737 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_001538_120 |
0.1008948264 |
- |
- |
PREDICTED: FAD-linked sulfhydryl oxidase ERV1 [Jatropha curcas] |
| 18 |
Hb_000531_020 |
0.1010153827 |
- |
- |
PREDICTED: uncharacterized protein At4g28440-like [Populus euphratica] |
| 19 |
Hb_002296_060 |
0.1033482028 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_001951_110 |
0.1045369992 |
- |
- |
PREDICTED: DNA-binding protein DDB_G0278111 [Jatropha curcas] |