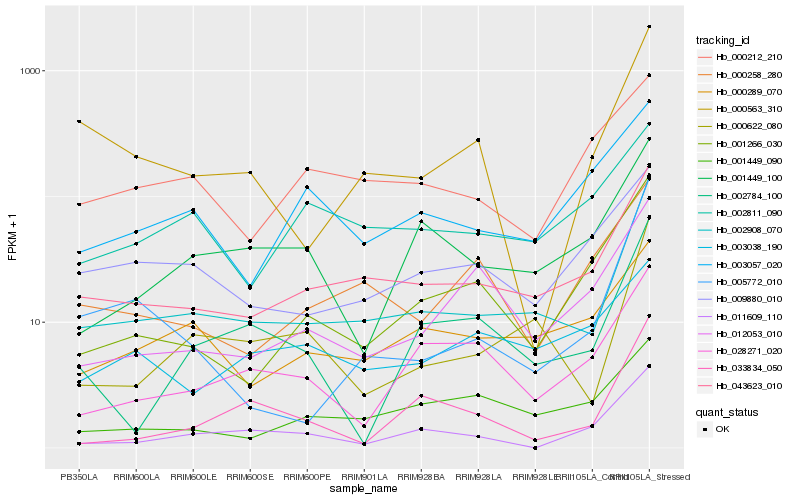
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002908_070 |
0.0 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 2 |
Hb_043623_010 |
0.1169745891 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 3 |
Hb_000622_080 |
0.1480619462 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 4 |
Hb_000289_070 |
0.1955967757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001449_100 |
0.2018009753 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 6 |
Hb_001266_030 |
0.2078703151 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_009880_010 |
0.2093301749 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 8 |
Hb_005772_010 |
0.2110310301 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 9 |
Hb_000563_310 |
0.2118242667 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 10 |
Hb_002784_100 |
0.2120360142 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 11 |
Hb_012053_010 |
0.2131254263 |
- |
- |
srpk, putative [Ricinus communis] |
| 12 |
Hb_001449_090 |
0.214893764 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_028271_020 |
0.215547984 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 14 |
Hb_003057_020 |
0.2161731733 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 15 |
Hb_002811_090 |
0.2191135602 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 16 |
Hb_000258_280 |
0.2204904363 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 17 |
Hb_011609_110 |
0.220971304 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 18 |
Hb_033834_050 |
0.2256664405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000212_210 |
0.228077981 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_003038_190 |
0.2306835783 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |