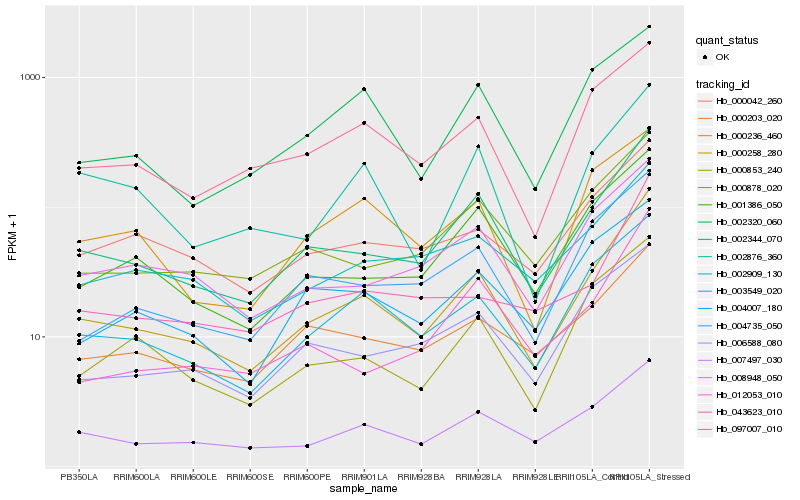
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_280 |
0.0 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 2 |
Hb_002344_070 |
0.0690106979 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 3 |
Hb_003549_020 |
0.1013366264 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007497_030 |
0.1021665407 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 5 |
Hb_001386_050 |
0.1208000076 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit gamma-like [Jatropha curcas] |
| 6 |
Hb_000878_020 |
0.1240196693 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 7 |
Hb_004735_050 |
0.1245543069 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 8 |
Hb_097007_010 |
0.1249846919 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 9 |
Hb_000853_240 |
0.1256636053 |
- |
- |
- |
| 10 |
Hb_008948_050 |
0.1286957653 |
- |
- |
hypothetical protein JCGZ_13339 [Jatropha curcas] |
| 11 |
Hb_002909_130 |
0.1366359116 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_006588_080 |
0.1368922858 |
- |
- |
hypothetical protein JCGZ_25746 [Jatropha curcas] |
| 13 |
Hb_043623_010 |
0.1373012825 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 14 |
Hb_000042_260 |
0.1373470725 |
- |
- |
PREDICTED: uncharacterized protein LOC105632814 [Jatropha curcas] |
| 15 |
Hb_000203_020 |
0.1389448349 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 16 |
Hb_004007_180 |
0.1397266985 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 17 |
Hb_000236_460 |
0.1397462553 |
- |
- |
PREDICTED: uncharacterized protein LOC105633119 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002876_360 |
0.1400449063 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 19 |
Hb_012053_010 |
0.1406511266 |
- |
- |
srpk, putative [Ricinus communis] |
| 20 |
Hb_002320_060 |
0.1427120423 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |