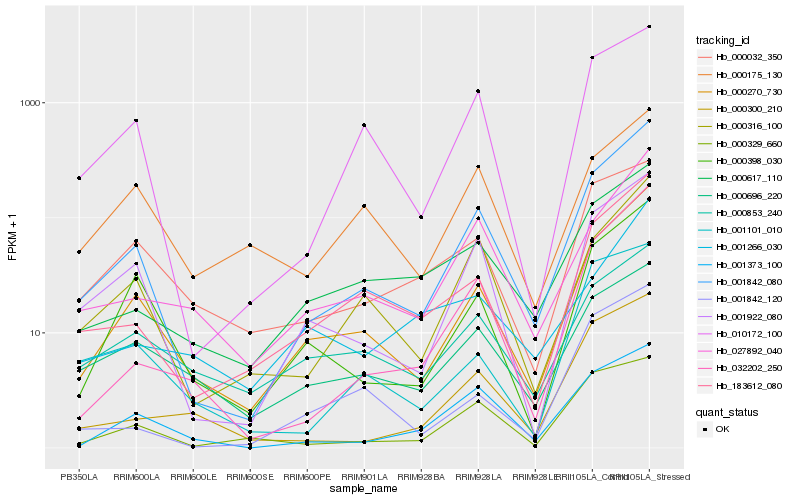
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_730 |
0.0 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 2 |
Hb_001842_080 |
0.0965544854 |
- |
- |
- |
| 3 |
Hb_000398_030 |
0.109450824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_183612_080 |
0.1307616732 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 5 |
Hb_001922_080 |
0.1428498901 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 6 |
Hb_000316_100 |
0.1464848711 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_027892_040 |
0.1475562443 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 8 |
Hb_000853_240 |
0.1588551612 |
- |
- |
- |
| 9 |
Hb_032202_250 |
0.1643087107 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 10 |
Hb_000617_110 |
0.16947982 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 11 |
Hb_010172_100 |
0.171161368 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 12 |
Hb_000696_220 |
0.1724263281 |
- |
- |
PREDICTED: uncharacterized protein LOC105647113 [Jatropha curcas] |
| 13 |
Hb_000032_350 |
0.1744389794 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001373_100 |
0.1790397302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001842_120 |
0.1817659474 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_001101_010 |
0.184579619 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 17 |
Hb_000175_130 |
0.1851933064 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 18 |
Hb_001266_030 |
0.1855550821 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000300_210 |
0.1867149982 |
- |
- |
gd2b, putative [Ricinus communis] |
| 20 |
Hb_000329_660 |
0.1870804797 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |