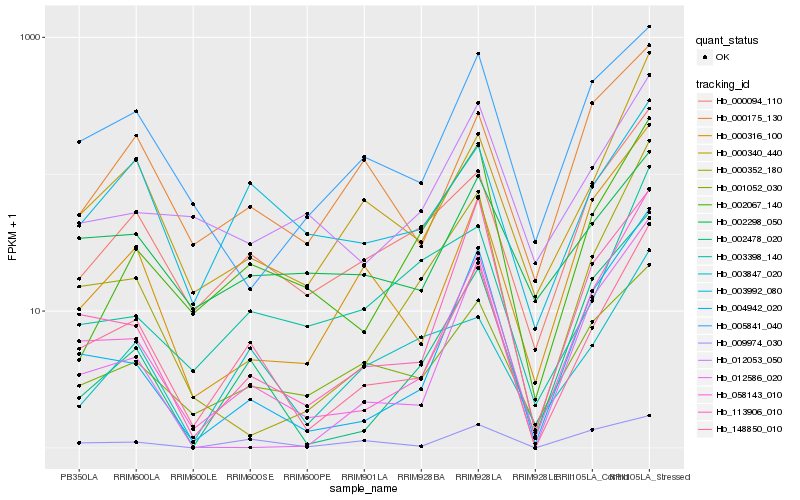
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148850_010 |
0.0 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 2 |
Hb_058143_010 |
0.1643533136 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 3 |
Hb_000094_110 |
0.1716866528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000340_440 |
0.1717613952 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 5 |
Hb_004942_020 |
0.1798020608 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 6 |
Hb_113906_010 |
0.1801983305 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 7 |
Hb_003992_080 |
0.1843447079 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 8 |
Hb_000352_180 |
0.1865314408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001052_030 |
0.1890280058 |
- |
- |
PREDICTED: protein TIC 20-IV, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002067_140 |
0.1913417218 |
- |
- |
- |
| 11 |
Hb_002478_020 |
0.1942800633 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 12 |
Hb_009974_030 |
0.1956660622 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_003847_020 |
0.2019434779 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_005841_040 |
0.2097700991 |
- |
- |
stress-induced hydrophobic peptide 1 [Hevea brasiliensis] |
| 15 |
Hb_000316_100 |
0.2100194581 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_002298_050 |
0.210638698 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 17 |
Hb_003398_140 |
0.2119209912 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_012053_050 |
0.2126146375 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 19 |
Hb_000175_130 |
0.2145932351 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 20 |
Hb_012586_020 |
0.2171998971 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |