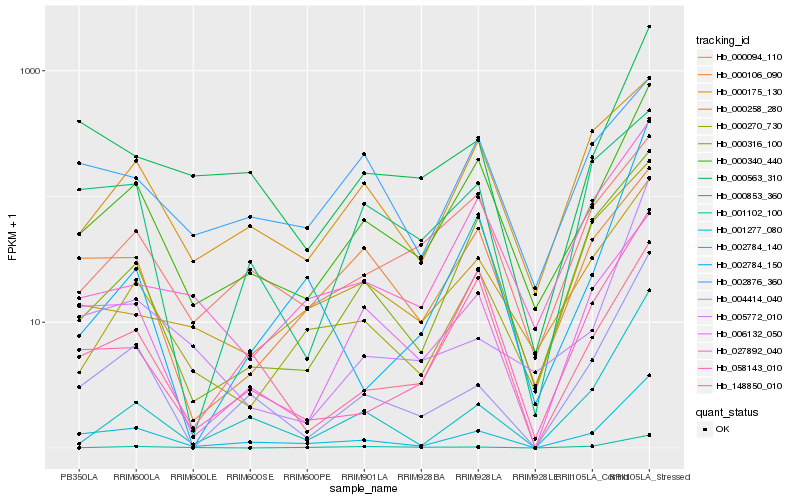
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002784_140 |
0.0 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 2 |
Hb_004414_040 |
0.1275276346 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_000340_440 |
0.1385918531 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 4 |
Hb_000563_310 |
0.1675656736 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 5 |
Hb_006132_050 |
0.1703272466 |
- |
- |
hypothetical protein EUTSA_v10028860mg [Eutrema salsugineum] |
| 6 |
Hb_001277_080 |
0.1963251193 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 7 |
Hb_000316_100 |
0.2002724267 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_005772_010 |
0.2011041307 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 9 |
Hb_058143_010 |
0.2090130825 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 10 |
Hb_000106_090 |
0.2113538187 |
- |
- |
PREDICTED: uncharacterized protein LOC105634795 [Jatropha curcas] |
| 11 |
Hb_000175_130 |
0.2145892259 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase At3g15890 [Jatropha curcas] |
| 12 |
Hb_001102_100 |
0.2153126696 |
- |
- |
PREDICTED: uncharacterized protein LOC105140429 isoform X2 [Populus euphratica] |
| 13 |
Hb_148850_010 |
0.2219801881 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 14 |
Hb_002876_360 |
0.2228355297 |
- |
- |
ubiquitin-conjugating enzyme E2, putative [Ricinus communis] |
| 15 |
Hb_000258_280 |
0.2236219806 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 16 |
Hb_000270_730 |
0.2242246008 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 17 |
Hb_000853_360 |
0.225567071 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 18 |
Hb_002784_150 |
0.226254532 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 19 |
Hb_027892_040 |
0.2271590058 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 20 |
Hb_000094_110 |
0.2278289093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |