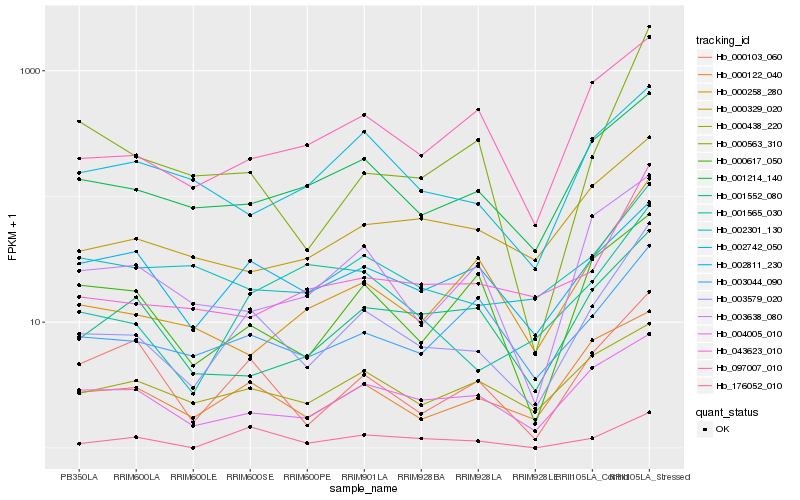
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003579_020 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 2 |
Hb_000122_040 |
0.1610154629 |
- |
- |
PREDICTED: uncharacterized protein LOC105649224 [Jatropha curcas] |
| 3 |
Hb_000103_060 |
0.1731138811 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 4 |
Hb_001214_140 |
0.1746809186 |
- |
- |
60S ribosomal protein L27a, putative [Ricinus communis] |
| 5 |
Hb_097007_010 |
0.1821938088 |
- |
- |
hypothetical protein CARUB_v10018269mg, partial [Capsella rubella] |
| 6 |
Hb_043623_010 |
0.1859179495 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 7 |
Hb_004005_010 |
0.1868499139 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 8 |
Hb_003044_090 |
0.1914061423 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 9 |
Hb_003638_080 |
0.19156208 |
- |
- |
hypothetical protein POPTR_0006s13340g [Populus trichocarpa] |
| 10 |
Hb_000617_050 |
0.1998072986 |
- |
- |
PREDICTED: uncharacterized protein LOC105647466 [Jatropha curcas] |
| 11 |
Hb_176052_010 |
0.199915555 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 12 |
Hb_000438_220 |
0.200068904 |
- |
- |
PREDICTED: uncharacterized protein LOC105649213 [Jatropha curcas] |
| 13 |
Hb_000563_310 |
0.2014607672 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3 [Hevea brasiliensis] |
| 14 |
Hb_002811_230 |
0.2023451023 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 15 |
Hb_001552_080 |
0.2036293533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002301_130 |
0.2038268784 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X3 [Populus euphratica] |
| 17 |
Hb_000329_020 |
0.2038685688 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 18 |
Hb_000258_280 |
0.2047846686 |
- |
- |
PREDICTED: probable ran guanine nucleotide release factor [Jatropha curcas] |
| 19 |
Hb_001565_030 |
0.2049755837 |
- |
- |
- |
| 20 |
Hb_002742_050 |
0.2057315236 |
- |
- |
hypothetical protein CICLE_v10002863mg [Citrus clementina] |