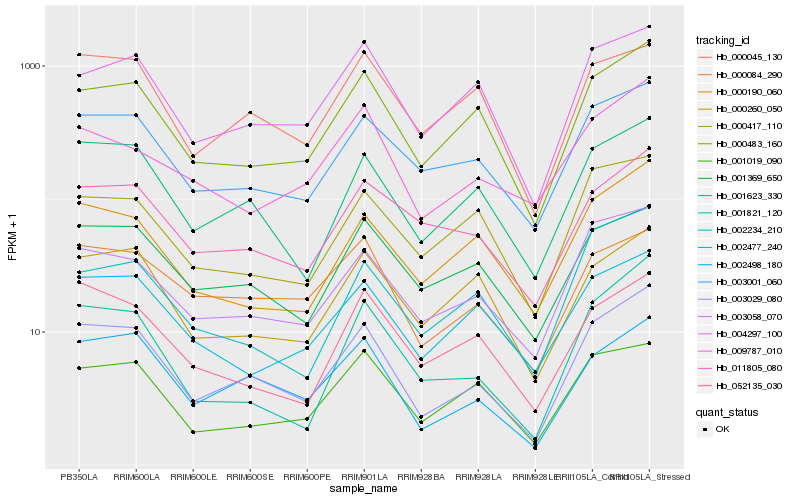
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003029_080 |
0.0 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 2 |
Hb_002498_180 |
0.095495621 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 3 |
Hb_001623_330 |
0.0956893189 |
- |
- |
PREDICTED: uncharacterized protein LOC105638463 [Jatropha curcas] |
| 4 |
Hb_003058_070 |
0.1004540361 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 5 |
Hb_000483_160 |
0.1020845221 |
- |
- |
PREDICTED: 60S ribosomal protein L34 [Jatropha curcas] |
| 6 |
Hb_003001_060 |
0.1115417449 |
- |
- |
wound-responsive family protein [Populus trichocarpa] |
| 7 |
Hb_001821_120 |
0.1153944899 |
- |
- |
PREDICTED: uncharacterized protein LOC105643956 [Jatropha curcas] |
| 8 |
Hb_000190_060 |
0.1206298408 |
- |
- |
PREDICTED: uncharacterized protein LOC105644688 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004297_100 |
0.1258917059 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P2B-like [Populus euphratica] |
| 10 |
Hb_011805_080 |
0.1286969718 |
- |
- |
PREDICTED: uncharacterized protein LOC100242357 [Vitis vinifera] |
| 11 |
Hb_002234_210 |
0.1309311972 |
- |
- |
hypothetical protein POPTR_0012s13380g [Populus trichocarpa] |
| 12 |
Hb_001369_650 |
0.1314525647 |
- |
- |
PREDICTED: chloride conductance regulatory protein ICln [Jatropha curcas] |
| 13 |
Hb_000084_290 |
0.1317036929 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 14 |
Hb_000260_050 |
0.1352005288 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 15 |
Hb_052135_030 |
0.1358848437 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 16 |
Hb_000417_110 |
0.1360503334 |
- |
- |
PREDICTED: uncharacterized protein LOC105649681 [Jatropha curcas] |
| 17 |
Hb_001019_090 |
0.1362198155 |
- |
- |
PREDICTED: uncharacterized protein LOC105639312 isoform X2 [Jatropha curcas] |
| 18 |
Hb_009787_010 |
0.1374106616 |
- |
- |
PREDICTED: 10 kDa chaperonin [Jatropha curcas] |
| 19 |
Hb_000045_130 |
0.1375302052 |
- |
- |
60S acidic ribosomal protein P0A [Hevea brasiliensis] |
| 20 |
Hb_002477_240 |
0.1378999857 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |