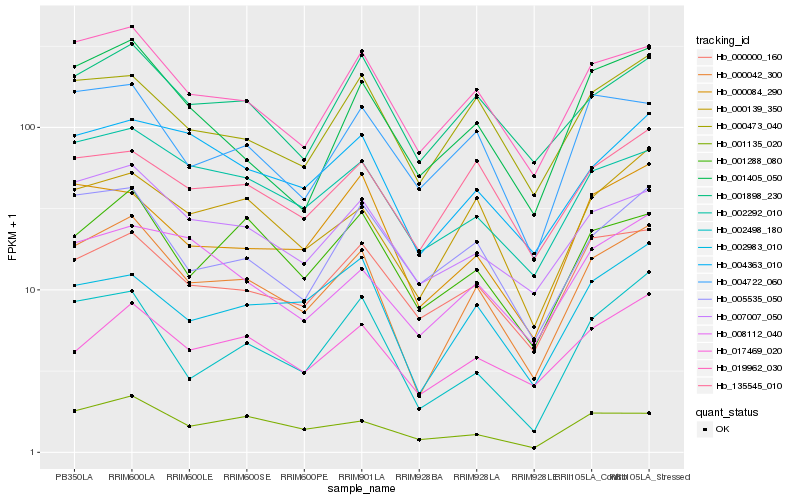
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_300 |
0.0 |
- |
- |
- |
| 2 |
Hb_019962_030 |
0.0934710808 |
- |
- |
PREDICTED: uncharacterized protein LOC105650622 [Jatropha curcas] |
| 3 |
Hb_007007_050 |
0.105488586 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 4 |
Hb_000139_350 |
0.1070872404 |
- |
- |
AtGCP3 interacting protein 1 [Theobroma cacao] |
| 5 |
Hb_002292_010 |
0.1082811942 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 6 |
Hb_004363_010 |
0.1123036086 |
- |
- |
hypothetical protein CISIN_1g0405721mg, partial [Citrus sinensis] |
| 7 |
Hb_001898_230 |
0.115450587 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000473_040 |
0.1182716818 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: RNA polymerase II transcriptional coactivator KIWI-like [Jatropha curcas] |
| 9 |
Hb_005535_050 |
0.1191849163 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_001288_080 |
0.1203869147 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 11 |
Hb_000000_160 |
0.1207049808 |
- |
- |
PREDICTED: membrin-11 [Jatropha curcas] |
| 12 |
Hb_002983_010 |
0.1213060867 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 13 |
Hb_002498_180 |
0.1213426005 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 14 |
Hb_001135_020 |
0.1220313986 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 15 |
Hb_000084_290 |
0.1229905631 |
- |
- |
mitochondrial 39S ribosomal protein L53 [Hevea brasiliensis] |
| 16 |
Hb_135545_010 |
0.1245365523 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_001405_050 |
0.1245878989 |
- |
- |
PREDICTED: uncharacterized protein LOC100500641 [Glycine max] |
| 18 |
Hb_017469_020 |
0.125153681 |
- |
- |
- |
| 19 |
Hb_008112_040 |
0.1261488551 |
- |
- |
PREDICTED: uncharacterized protein LOC105637336 [Jatropha curcas] |
| 20 |
Hb_004722_060 |
0.126900934 |
- |
- |
PREDICTED: protein CLP1 homolog [Jatropha curcas] |