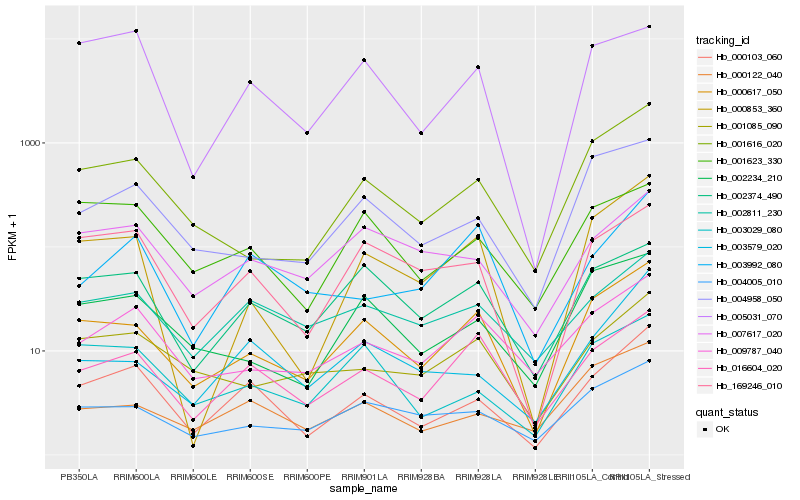
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 2 |
Hb_000617_050 |
0.1687281067 |
- |
- |
PREDICTED: uncharacterized protein LOC105647466 [Jatropha curcas] |
| 3 |
Hb_169246_010 |
0.1690053237 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003579_020 |
0.1731138811 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 5 |
Hb_002811_230 |
0.1766530986 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 6 |
Hb_003029_080 |
0.1802652514 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 7 |
Hb_000122_040 |
0.1803792953 |
- |
- |
PREDICTED: uncharacterized protein LOC105649224 [Jatropha curcas] |
| 8 |
Hb_016604_020 |
0.1822229952 |
- |
- |
- |
| 9 |
Hb_007617_020 |
0.1908326446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001616_020 |
0.1908666017 |
- |
- |
PREDICTED: wound-induced basic protein [Jatropha curcas] |
| 11 |
Hb_005031_070 |
0.19462627 |
- |
- |
RecName: Full=Major latex allergen Hev b 5; AltName: Allergen=Hev b 5 [Hevea brasiliensis] |
| 12 |
Hb_004005_010 |
0.1950119021 |
- |
- |
PREDICTED: uncharacterized protein LOC102623333 isoform X2 [Citrus sinensis] |
| 13 |
Hb_001623_330 |
0.1950154921 |
- |
- |
PREDICTED: uncharacterized protein LOC105638463 [Jatropha curcas] |
| 14 |
Hb_000853_360 |
0.196963006 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 15 |
Hb_009787_040 |
0.1988588184 |
desease resistance |
Gene Name: AAA_17 |
PREDICTED: uncharacterized protein LOC105135462 isoform X2 [Populus euphratica] |
| 16 |
Hb_003992_080 |
0.2002679003 |
- |
- |
PREDICTED: protein LURP-one-related 5 [Jatropha curcas] |
| 17 |
Hb_002374_490 |
0.2011572739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004958_050 |
0.2012359855 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 19 |
Hb_002234_210 |
0.205740381 |
- |
- |
hypothetical protein POPTR_0012s13380g [Populus trichocarpa] |
| 20 |
Hb_001085_090 |
0.2059750411 |
- |
- |
PREDICTED: uncharacterized protein LOC105634186 [Jatropha curcas] |