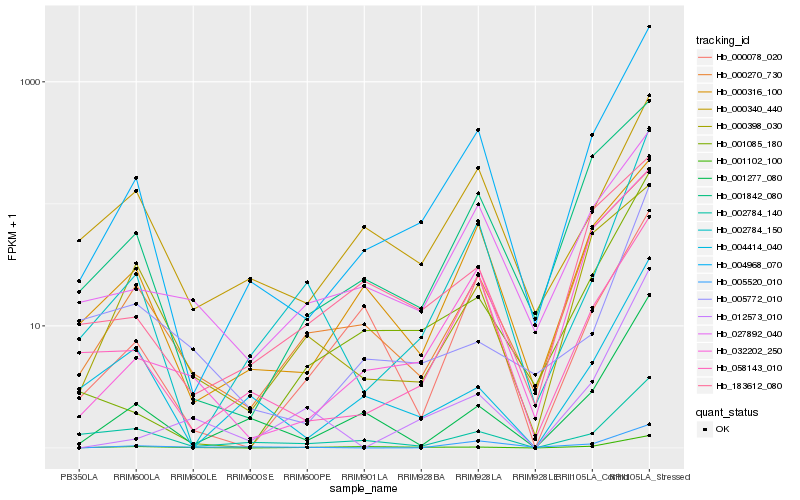
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_080 |
0.0 |
- |
- |
PREDICTED: vegetative cell wall protein gp1 [Jatropha curcas] |
| 2 |
Hb_004414_040 |
0.1622583569 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_004968_070 |
0.1723663279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002784_140 |
0.1963251193 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 5 |
Hb_001102_100 |
0.2064923643 |
- |
- |
PREDICTED: uncharacterized protein LOC105140429 isoform X2 [Populus euphratica] |
| 6 |
Hb_000270_730 |
0.2102117445 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa-like [Jatropha curcas] |
| 7 |
Hb_002784_150 |
0.2281339505 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 8 |
Hb_000340_440 |
0.230820871 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 9 |
Hb_000316_100 |
0.231490361 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000078_020 |
0.2367628667 |
- |
- |
- |
| 11 |
Hb_001085_180 |
0.2379791809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_183612_080 |
0.2386414767 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 13 |
Hb_001842_080 |
0.2406190745 |
- |
- |
- |
| 14 |
Hb_005772_010 |
0.2417617982 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 15 |
Hb_005520_010 |
0.2422065203 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 16 |
Hb_027892_040 |
0.2490776119 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 17 |
Hb_032202_250 |
0.2521971177 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 18 |
Hb_000398_030 |
0.2600988813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_058143_010 |
0.2613443036 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 20 |
Hb_012573_010 |
0.2644314976 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |