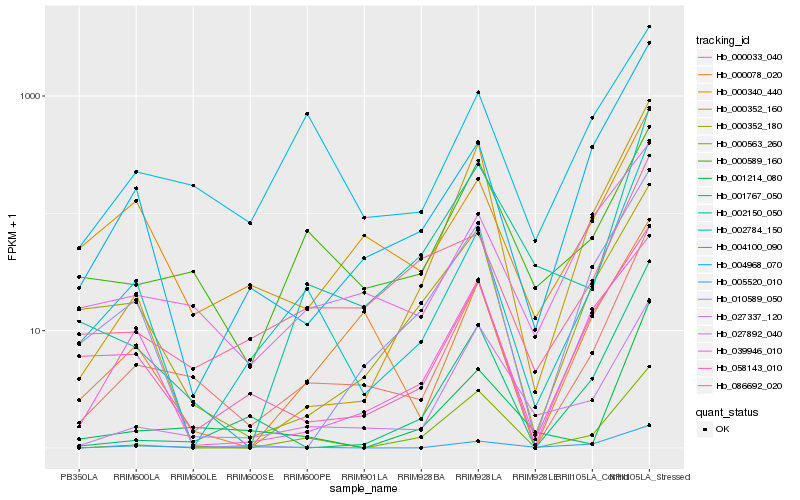
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000033_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_027337_120 |
0.1771741013 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 3 |
Hb_002784_150 |
0.179685295 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 4 |
Hb_027892_040 |
0.1808018624 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 5 |
Hb_000589_160 |
0.1832608803 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 6 |
Hb_058143_010 |
0.1878406483 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 7 |
Hb_000078_020 |
0.1893484739 |
- |
- |
- |
| 8 |
Hb_004100_090 |
0.1930266687 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |
| 9 |
Hb_005520_010 |
0.194017747 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 10 |
Hb_010589_050 |
0.1952934874 |
- |
- |
- |
| 11 |
Hb_000352_160 |
0.1971193584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_086692_020 |
0.2011169578 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004968_070 |
0.2045542628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000352_180 |
0.2047517131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000340_440 |
0.2057093709 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 16 |
Hb_001767_050 |
0.2063015969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_039946_010 |
0.2068852852 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 18 |
Hb_002150_050 |
0.209257364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000563_260 |
0.2095868035 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 20 |
Hb_001214_080 |
0.2118904638 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |