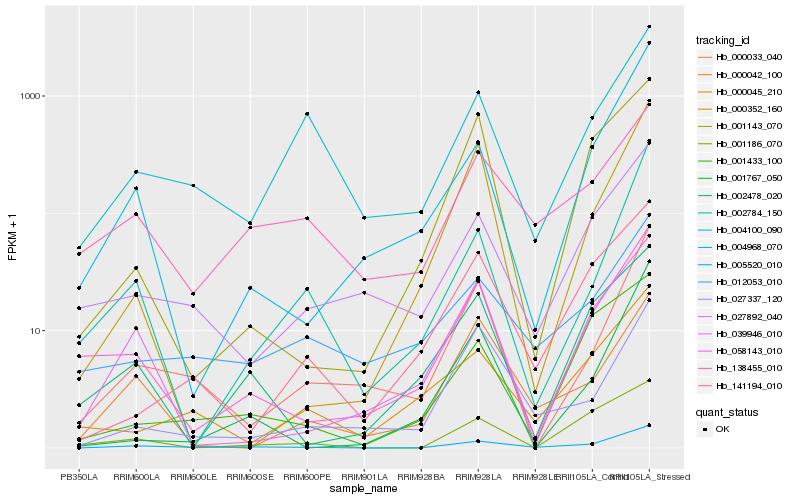
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005520_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 2 |
Hb_004968_070 |
0.1913314734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000033_040 |
0.194017747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_058143_010 |
0.1972030666 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 5 |
Hb_002784_150 |
0.1990871566 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 6 |
Hb_027892_040 |
0.2022585156 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 7 |
Hb_141194_010 |
0.2103594296 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 8 |
Hb_001767_050 |
0.2127189465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001186_070 |
0.2136060706 |
- |
- |
PREDICTED: F-box protein FBW2-like [Gossypium raimondii] |
| 10 |
Hb_001433_100 |
0.2142876681 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004100_090 |
0.2146948249 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |
| 12 |
Hb_039946_010 |
0.2181903036 |
- |
- |
hypothetical protein POPTR_0018s09200g [Populus trichocarpa] |
| 13 |
Hb_027337_120 |
0.2189332847 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 14 |
Hb_002478_020 |
0.2191489364 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |
| 15 |
Hb_138455_010 |
0.219989993 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_012053_010 |
0.2233354921 |
- |
- |
srpk, putative [Ricinus communis] |
| 17 |
Hb_000045_210 |
0.2241234131 |
- |
- |
- |
| 18 |
Hb_000042_100 |
0.2276592233 |
- |
- |
PREDICTED: uncharacterized protein LOC104448410 [Eucalyptus grandis] |
| 19 |
Hb_000352_160 |
0.2278880477 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001143_070 |
0.2289695613 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |