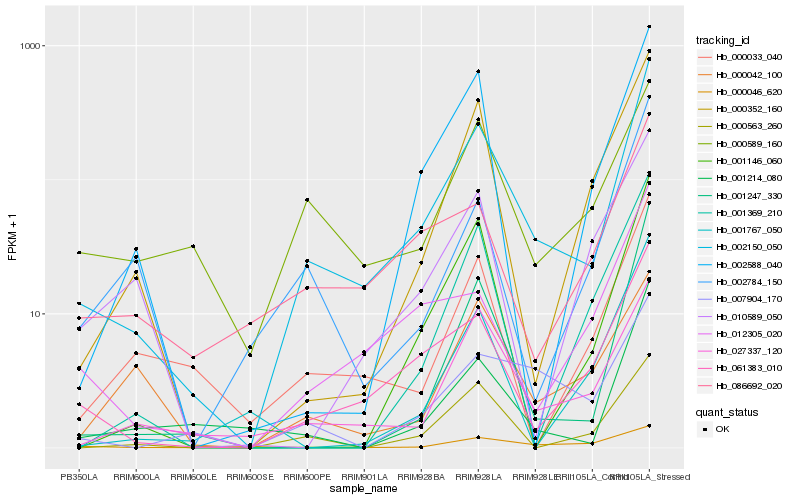
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_061383_010 |
0.1606890214 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 3 |
Hb_027337_120 |
0.1741501227 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 4 |
Hb_001247_330 |
0.1862329132 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 5 |
Hb_001214_080 |
0.1930668372 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 6 |
Hb_000563_260 |
0.1955923778 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 7 |
Hb_001146_060 |
0.2013151738 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 8 |
Hb_002588_040 |
0.202338084 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 9 |
Hb_086692_020 |
0.2029193587 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_012305_020 |
0.206109338 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 11 |
Hb_000352_160 |
0.2066129973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000033_040 |
0.209257364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000589_160 |
0.21529017 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 14 |
Hb_007904_170 |
0.2174304371 |
- |
- |
PREDICTED: anther-specific protein BCP1 [Jatropha curcas] |
| 15 |
Hb_002784_150 |
0.2215666307 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 16 |
Hb_001369_210 |
0.2231669157 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 17 |
Hb_001767_050 |
0.2260265445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000046_620 |
0.2352089682 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 19 |
Hb_000042_100 |
0.2367732972 |
- |
- |
PREDICTED: uncharacterized protein LOC104448410 [Eucalyptus grandis] |
| 20 |
Hb_010589_050 |
0.2373248424 |
- |
- |
- |