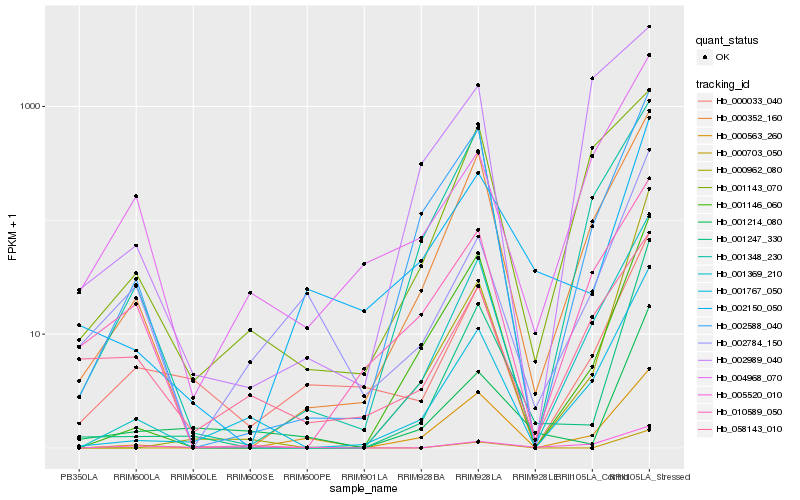
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001767_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001369_210 |
0.127783927 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 3 |
Hb_000352_160 |
0.142714195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000962_080 |
0.1612806162 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 5 |
Hb_001146_060 |
0.1683706687 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 6 |
Hb_002588_040 |
0.1707742981 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 7 |
Hb_001247_330 |
0.1730458348 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 8 |
Hb_001348_230 |
0.1864424382 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_004968_070 |
0.1930046296 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001143_070 |
0.2061686368 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000033_040 |
0.2063015969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010589_050 |
0.2101087649 |
- |
- |
- |
| 13 |
Hb_058143_010 |
0.2113550705 |
- |
- |
hypothetical protein JCGZ_01992 [Jatropha curcas] |
| 14 |
Hb_005520_010 |
0.2127189465 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 15 |
Hb_000563_260 |
0.2133214633 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 16 |
Hb_001214_080 |
0.2191769817 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 17 |
Hb_000703_050 |
0.2199348991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002784_150 |
0.220516681 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 19 |
Hb_002989_040 |
0.2253847555 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 20 |
Hb_002150_050 |
0.2260265445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |