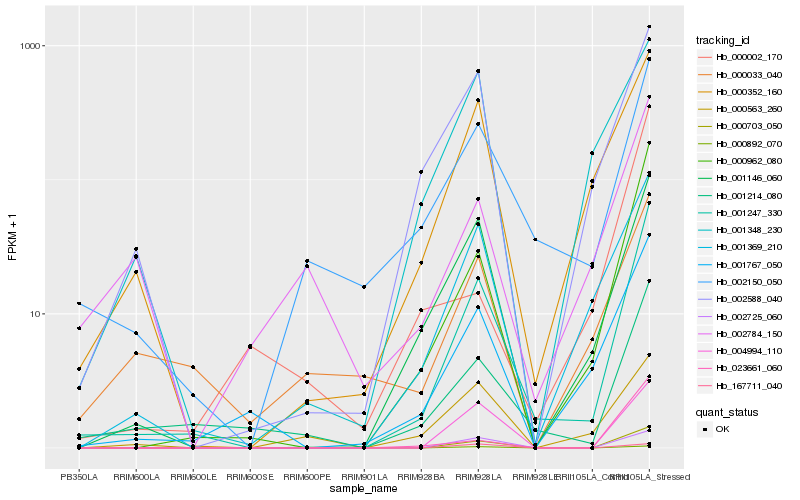
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004994_110 |
0.1193125748 |
- |
- |
- |
| 3 |
Hb_002725_060 |
0.122393099 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 4 |
Hb_001247_330 |
0.1264037803 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 5 |
Hb_000962_080 |
0.1614595218 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 6 |
Hb_001146_060 |
0.2044518325 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 7 |
Hb_001767_050 |
0.2199348991 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001369_210 |
0.220856398 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 9 |
Hb_167711_040 |
0.226433891 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 10 |
Hb_023661_060 |
0.2299076275 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_000352_160 |
0.2396041987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002588_040 |
0.2463999201 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 13 |
Hb_001214_080 |
0.2491913423 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 14 |
Hb_002150_050 |
0.2612494685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000563_260 |
0.2703034476 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 16 |
Hb_000892_070 |
0.2720118403 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_001348_230 |
0.2851143337 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000033_040 |
0.2972562139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002784_150 |
0.3019303801 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 20 |
Hb_000002_170 |
0.3104887595 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |