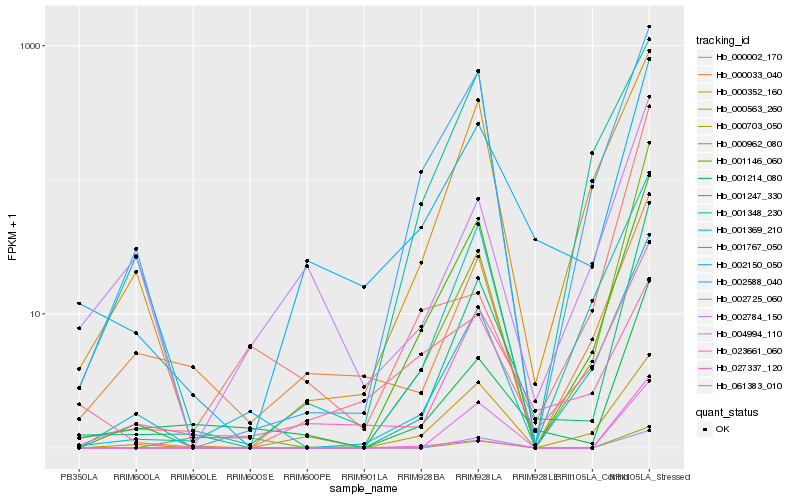
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001247_330 |
0.0 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 2 |
Hb_000703_050 |
0.1264037803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000962_080 |
0.1288992591 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 4 |
Hb_001767_050 |
0.1730458348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001146_060 |
0.1734362014 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 6 |
Hb_001214_080 |
0.1748311202 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 7 |
Hb_001369_210 |
0.1846465567 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 8 |
Hb_004994_110 |
0.1854347747 |
- |
- |
- |
| 9 |
Hb_002150_050 |
0.1862329132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002725_060 |
0.1876965466 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 11 |
Hb_000352_160 |
0.1916229209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002588_040 |
0.2033837159 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 13 |
Hb_023661_060 |
0.2250628639 |
- |
- |
PREDICTED: uncharacterized protein LOC104886268, partial [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000563_260 |
0.2407293359 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 15 |
Hb_000033_040 |
0.2463780344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001348_230 |
0.2475875798 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_002784_150 |
0.2485792167 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 18 |
Hb_027337_120 |
0.2551206702 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 19 |
Hb_061383_010 |
0.2640761614 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 20 |
Hb_000002_170 |
0.2666728387 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |