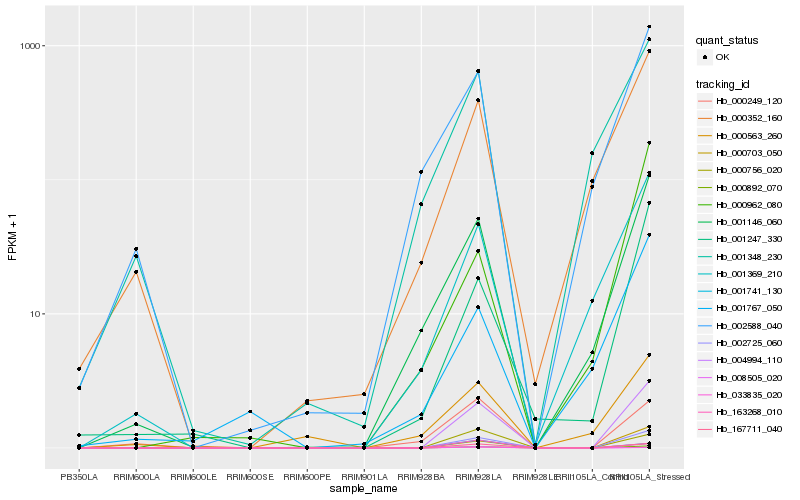
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_002725_060 |
0.0031014403 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 3 |
Hb_167711_040 |
0.1083361769 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 4 |
Hb_000703_050 |
0.1193125748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001247_330 |
0.1854347747 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 6 |
Hb_001146_060 |
0.1871272687 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 7 |
Hb_000756_020 |
0.2033102437 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 8 |
Hb_001369_210 |
0.2204630918 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_26269 [Jatropha curcas] |
| 9 |
Hb_000892_070 |
0.2313980993 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_002588_040 |
0.2342074218 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 11 |
Hb_000352_160 |
0.2342916496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000249_120 |
0.2345624299 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 13 |
Hb_163268_010 |
0.2401983731 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 14 |
Hb_001741_130 |
0.2403603433 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 15 |
Hb_000563_260 |
0.2477703706 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 16 |
Hb_000962_080 |
0.2532776783 |
- |
- |
PREDICTED: uncharacterized protein LOC105131773 [Populus euphratica] |
| 17 |
Hb_001767_050 |
0.2554879663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001348_230 |
0.2559762903 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_008505_020 |
0.2590128335 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 20 |
Hb_033835_020 |
0.2600398128 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |