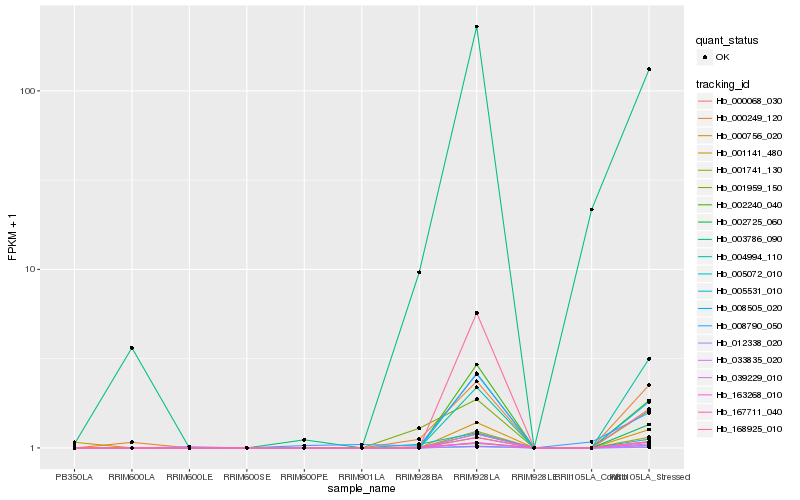
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005531_010 |
0.0 |
- |
- |
hypothetical protein CICLE_v10033008mg [Citrus clementina] |
| 2 |
Hb_033835_020 |
0.0073197596 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_008505_020 |
0.0083719662 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_002240_040 |
0.0269288752 |
- |
- |
- |
| 5 |
Hb_001741_130 |
0.027442861 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 6 |
Hb_163268_010 |
0.027608139 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 7 |
Hb_000756_020 |
0.065109017 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 8 |
Hb_039229_010 |
0.111738201 |
- |
- |
- |
| 9 |
Hb_167711_040 |
0.16045299 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 10 |
Hb_003786_090 |
0.2072793634 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 11 |
Hb_008790_050 |
0.2105538053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_168925_010 |
0.2181711919 |
- |
- |
- |
| 13 |
Hb_000249_120 |
0.2247611582 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 14 |
Hb_000068_030 |
0.2540669726 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 15 |
Hb_005072_010 |
0.2589086988 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 16 |
Hb_002725_060 |
0.2641533675 |
- |
- |
PREDICTED: uncharacterized protein LOC105644961 [Jatropha curcas] |
| 17 |
Hb_012338_020 |
0.2656020883 |
- |
- |
- |
| 18 |
Hb_004994_110 |
0.267177783 |
- |
- |
- |
| 19 |
Hb_001141_480 |
0.2968751175 |
- |
- |
- |
| 20 |
Hb_001959_150 |
0.3013319035 |
- |
- |
- |