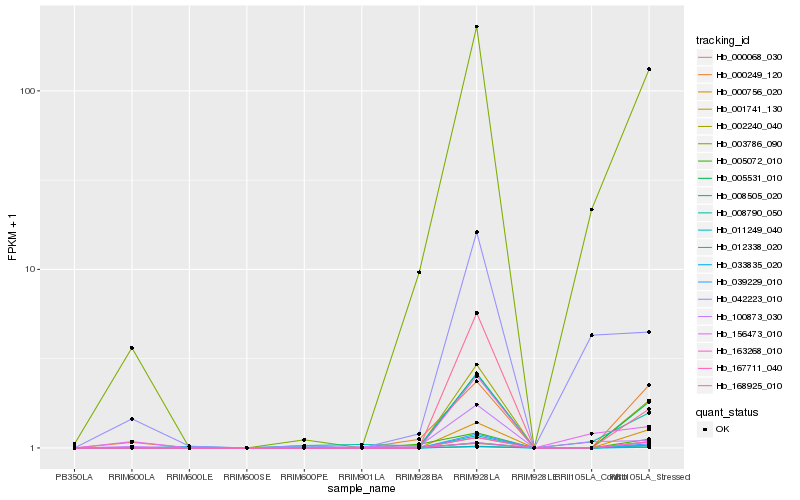
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039229_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002240_040 |
0.0849386188 |
- |
- |
- |
| 3 |
Hb_168925_010 |
0.1081938147 |
- |
- |
- |
| 4 |
Hb_005531_010 |
0.111738201 |
- |
- |
hypothetical protein CICLE_v10033008mg [Citrus clementina] |
| 5 |
Hb_033835_020 |
0.119010766 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_008505_020 |
0.1200557834 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 7 |
Hb_001741_130 |
0.1389793093 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Cucumis sativus] |
| 8 |
Hb_163268_010 |
0.1391431721 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 9 |
Hb_000756_020 |
0.1762653982 |
- |
- |
BnaA05g19440D [Brassica napus] |
| 10 |
Hb_008790_050 |
0.2063413424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003786_090 |
0.2464730354 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 12 |
Hb_011249_040 |
0.2594996216 |
- |
- |
PREDICTED: acetylajmalan esterase-like [Populus euphratica] |
| 13 |
Hb_167711_040 |
0.2701911958 |
- |
- |
PREDICTED: cucumisin-like [Jatropha curcas] |
| 14 |
Hb_156473_010 |
0.2706066035 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 15 |
Hb_000068_030 |
0.2813953396 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 16 |
Hb_005072_010 |
0.2883929352 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 17 |
Hb_012338_020 |
0.292185565 |
- |
- |
- |
| 18 |
Hb_000249_120 |
0.2985300894 |
- |
- |
PREDICTED: uncharacterized protein LOC104613364 [Nelumbo nucifera] |
| 19 |
Hb_100873_030 |
0.2996247907 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 20 |
Hb_042223_010 |
0.3087940147 |
- |
- |
Protein Z, putative [Ricinus communis] |