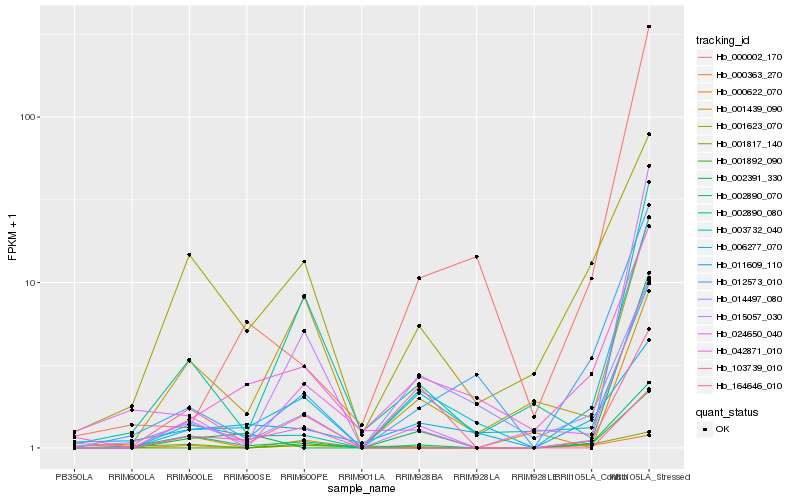
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002890_070 |
0.0 |
- |
- |
ATP-dependent RNA helicase family protein [Populus trichocarpa] |
| 2 |
Hb_001623_070 |
0.1979130998 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 3 |
Hb_006277_070 |
0.2149382125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012573_010 |
0.2279818241 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 5 |
Hb_002890_080 |
0.2281975084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003732_040 |
0.2504600758 |
- |
- |
putative esterase family protein [Populus trichocarpa] |
| 7 |
Hb_001439_090 |
0.252362669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_015057_030 |
0.2538890173 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52, partial [Jatropha curcas] |
| 9 |
Hb_002391_330 |
0.2577184436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_024650_040 |
0.2629628061 |
- |
- |
PREDICTED: josephin-like protein [Jatropha curcas] |
| 11 |
Hb_042871_010 |
0.2692737646 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 12 |
Hb_001892_090 |
0.2760372061 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_011609_110 |
0.2761965061 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 14 |
Hb_103739_010 |
0.2764838999 |
- |
- |
sodium/hydrogen exchanger, putative [Ricinus communis] |
| 15 |
Hb_164646_010 |
0.2765211775 |
- |
- |
PREDICTED: acidic endochitinase-like [Pyrus x bretschneideri] |
| 16 |
Hb_000363_270 |
0.2828640643 |
- |
- |
hypothetical protein PRUPE_ppa024846mg [Prunus persica] |
| 17 |
Hb_001817_140 |
0.2866631469 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_014497_080 |
0.287323872 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 19 |
Hb_000002_170 |
0.2902350427 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |
| 20 |
Hb_000622_070 |
0.2960008863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |