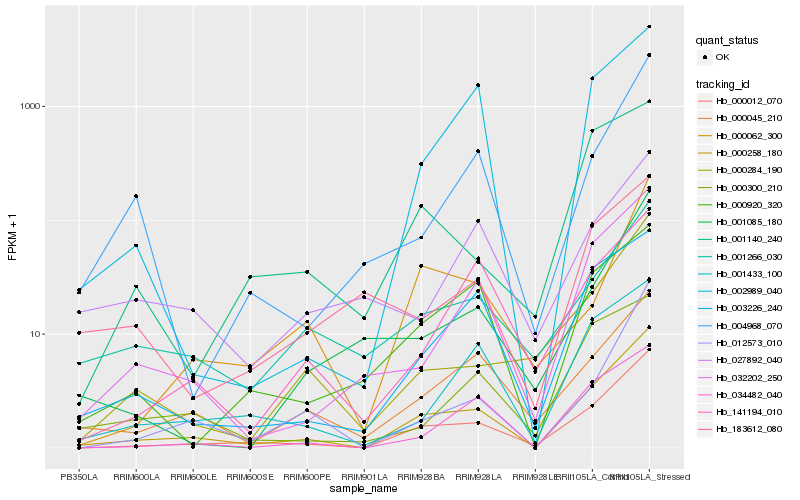
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_180 |
0.0 |
- |
- |
PREDICTED: glutathione S-transferase U10-like [Vitis vinifera] |
| 2 |
Hb_000012_070 |
0.088995292 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_032202_250 |
0.1623303032 |
- |
- |
PREDICTED: F-box protein PP2-A13 [Jatropha curcas] |
| 4 |
Hb_000045_210 |
0.1714765665 |
- |
- |
- |
| 5 |
Hb_034482_040 |
0.1734641119 |
- |
- |
aspartate kinase, partial [Francisella tularensis] |
| 6 |
Hb_003226_240 |
0.1751841203 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 7 |
Hb_001433_100 |
0.1771688492 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_012573_010 |
0.1807148985 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 9 |
Hb_002989_040 |
0.1856841266 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 10 |
Hb_000920_320 |
0.1909685067 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 11 |
Hb_001266_030 |
0.1955420061 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000284_190 |
0.1984733082 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_004968_070 |
0.1985111152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001085_180 |
0.2014089017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000300_210 |
0.205693656 |
- |
- |
gd2b, putative [Ricinus communis] |
| 16 |
Hb_000062_300 |
0.2085489835 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 17 |
Hb_141194_010 |
0.2087520685 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR 1-like isoform X2 [Populus euphratica] |
| 18 |
Hb_001140_240 |
0.215573636 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_183612_080 |
0.217797512 |
- |
- |
hypothetical protein POPTR_0001s00500g [Populus trichocarpa] |
| 20 |
Hb_027892_040 |
0.220912214 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |