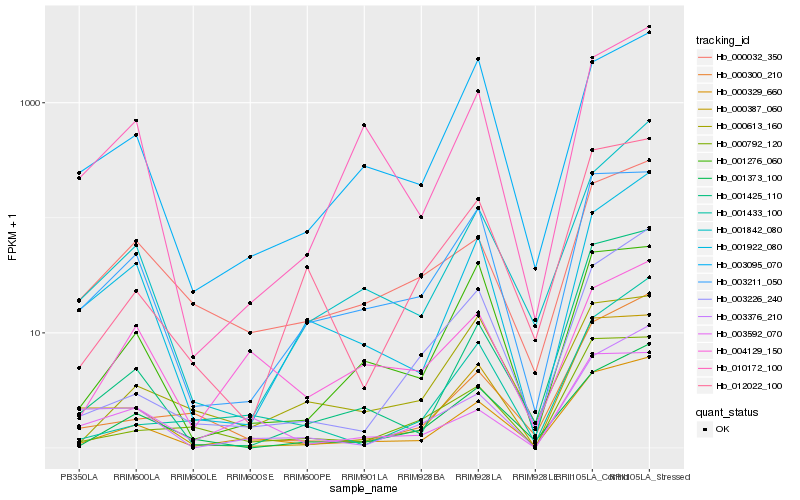
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_660 |
0.0 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 2 |
Hb_003211_050 |
0.1392212543 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 3 |
Hb_001373_100 |
0.1457757051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000387_060 |
0.1491333295 |
- |
- |
- |
| 5 |
Hb_000300_210 |
0.1566421817 |
- |
- |
gd2b, putative [Ricinus communis] |
| 6 |
Hb_003226_240 |
0.15700735 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 7 |
Hb_001276_060 |
0.1570174521 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 8 |
Hb_000032_350 |
0.158089934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001433_100 |
0.1581408956 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001922_080 |
0.1600836677 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 11 |
Hb_003095_070 |
0.1605307987 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 12 |
Hb_000792_120 |
0.1606299333 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001842_080 |
0.1619485885 |
- |
- |
- |
| 14 |
Hb_012022_100 |
0.1622346489 |
- |
- |
CPRD49 family protein [Populus trichocarpa] |
| 15 |
Hb_001425_110 |
0.1637771639 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 16 |
Hb_010172_100 |
0.1664435042 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Populus euphratica] |
| 17 |
Hb_003376_210 |
0.1706822799 |
- |
- |
- |
| 18 |
Hb_003592_070 |
0.171343299 |
- |
- |
- |
| 19 |
Hb_000613_160 |
0.1733030392 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 20 |
Hb_004129_150 |
0.1733533921 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |