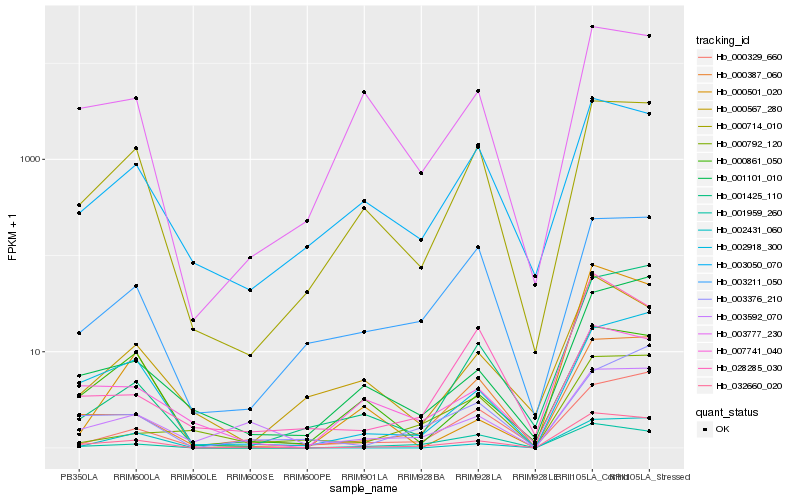
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032660_020 |
0.0 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 2 |
Hb_000387_060 |
0.140305122 |
- |
- |
- |
| 3 |
Hb_000714_010 |
0.1606394896 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 4 |
Hb_003592_070 |
0.1709504292 |
- |
- |
- |
| 5 |
Hb_001101_010 |
0.1742010371 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 6 |
Hb_002918_300 |
0.1757125109 |
- |
- |
- |
| 7 |
Hb_003050_070 |
0.1800116909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002431_060 |
0.1909907144 |
- |
- |
- |
| 9 |
Hb_003777_230 |
0.191001471 |
- |
- |
PREDICTED: abscisic stress-ripening protein 1-like [Citrus sinensis] |
| 10 |
Hb_001425_110 |
0.1922650323 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 11 |
Hb_007741_040 |
0.1924714809 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105643153 [Jatropha curcas] |
| 12 |
Hb_000329_660 |
0.1957820221 |
- |
- |
PREDICTED: carotenoid cleavage dioxygenase 7, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003211_050 |
0.19841606 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 14 |
Hb_028285_030 |
0.2001605965 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 1-like [Elaeis guineensis] |
| 15 |
Hb_000567_280 |
0.2083187934 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Populus euphratica] |
| 16 |
Hb_000501_020 |
0.2094551525 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 17 |
Hb_003376_210 |
0.2095657604 |
- |
- |
- |
| 18 |
Hb_001959_260 |
0.210303181 |
- |
- |
- |
| 19 |
Hb_000861_050 |
0.2156168625 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 20 |
Hb_000792_120 |
0.2172839698 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 1 isoform X2 [Jatropha curcas] |