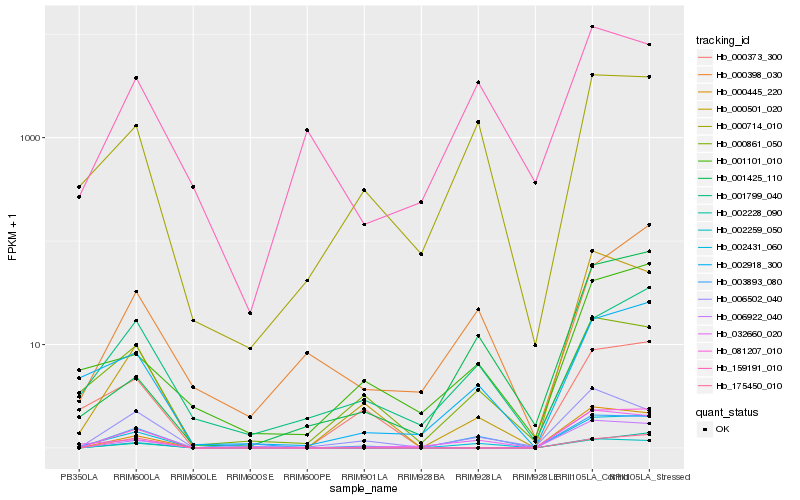
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002431_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_081207_010 |
0.1653045888 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 3 |
Hb_002918_300 |
0.1714974495 |
- |
- |
- |
| 4 |
Hb_002259_050 |
0.1863982046 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_032660_020 |
0.1909907144 |
- |
- |
PREDICTED: F-box protein At5g49610-like [Jatropha curcas] |
| 6 |
Hb_000445_220 |
0.1955447052 |
- |
- |
- |
| 7 |
Hb_002228_090 |
0.199502928 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 8 |
Hb_003893_080 |
0.2001016159 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006922_040 |
0.2060197758 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000714_010 |
0.2107413532 |
- |
- |
PREDICTED: salt stress-induced hydrophobic peptide ESI3 [Vitis vinifera] |
| 11 |
Hb_175450_010 |
0.216080547 |
- |
- |
- |
| 12 |
Hb_000861_050 |
0.2173246781 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000501_020 |
0.2253005923 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 14 |
Hb_001799_040 |
0.2307815288 |
- |
- |
- |
| 15 |
Hb_006502_040 |
0.2370768595 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 16 |
Hb_001101_010 |
0.2384609649 |
- |
- |
hypothetical protein POPTR_0004s18730g [Populus trichocarpa] |
| 17 |
Hb_001425_110 |
0.2437613713 |
- |
- |
hypothetical protein Csa_6G106200 [Cucumis sativus] |
| 18 |
Hb_000373_300 |
0.247065903 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 19 |
Hb_000398_030 |
0.24784901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_159191_010 |
0.2544482229 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |