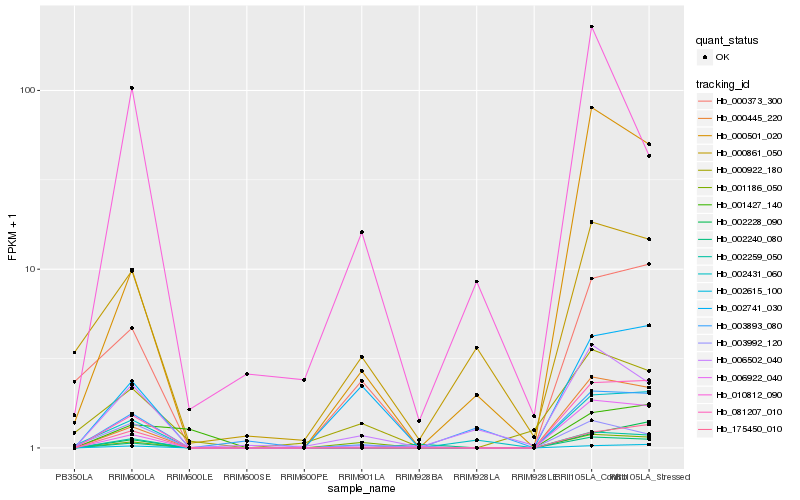
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002259_050 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_081207_010 |
0.0963883729 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 3 |
Hb_175450_010 |
0.1389636353 |
- |
- |
- |
| 4 |
Hb_000445_220 |
0.1496461217 |
- |
- |
- |
| 5 |
Hb_006922_040 |
0.1682308742 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002431_060 |
0.1863982046 |
- |
- |
- |
| 7 |
Hb_002228_090 |
0.1921463819 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 8 |
Hb_000501_020 |
0.2365652624 |
- |
- |
PREDICTED: uncharacterized protein LOC100250168 [Vitis vinifera] |
| 9 |
Hb_006502_040 |
0.2368996521 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 10 |
Hb_002615_100 |
0.2422514038 |
- |
- |
- |
| 11 |
Hb_003992_120 |
0.2478992431 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 12 |
Hb_002240_080 |
0.264471628 |
- |
- |
- |
| 13 |
Hb_000373_300 |
0.2648683975 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 14 |
Hb_003893_080 |
0.2746681966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002741_030 |
0.2798557932 |
- |
- |
- |
| 16 |
Hb_001186_050 |
0.283561005 |
- |
- |
- |
| 17 |
Hb_001427_140 |
0.2881997759 |
- |
- |
PREDICTED: probable phospholipase A2 homolog 1 [Jatropha curcas] |
| 18 |
Hb_000922_180 |
0.2913258476 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 19 |
Hb_010812_090 |
0.2961473627 |
- |
- |
PREDICTED: serine/threonine-protein kinase AtPK2/AtPK19-like [Jatropha curcas] |
| 20 |
Hb_000861_050 |
0.2969773732 |
- |
- |
PREDICTED: transcription elongation factor SPT6-like isoform X1 [Populus euphratica] |