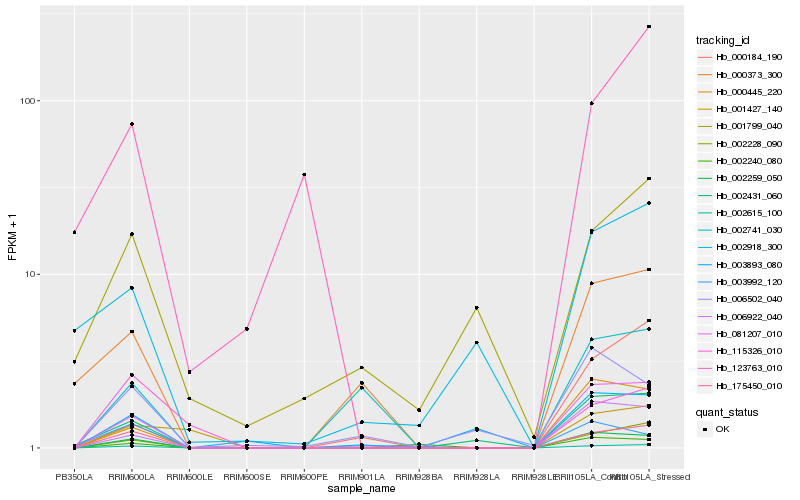
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175450_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002259_050 |
0.1389636353 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_002228_090 |
0.1534125535 |
- |
- |
hypothetical protein POPTR_0006s23560g [Populus trichocarpa] |
| 4 |
Hb_081207_010 |
0.1581639376 |
- |
- |
hypothetical protein JCGZ_17195 [Jatropha curcas] |
| 5 |
Hb_002615_100 |
0.2117126578 |
- |
- |
- |
| 6 |
Hb_002431_060 |
0.216080547 |
- |
- |
- |
| 7 |
Hb_000445_220 |
0.2461223412 |
- |
- |
- |
| 8 |
Hb_006922_040 |
0.2504648704 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_115326_010 |
0.2511721818 |
- |
- |
- |
| 10 |
Hb_000373_300 |
0.2764902159 |
- |
- |
hypothetical protein F775_20382 [Aegilops tauschii] |
| 11 |
Hb_001427_140 |
0.2871752243 |
- |
- |
PREDICTED: probable phospholipase A2 homolog 1 [Jatropha curcas] |
| 12 |
Hb_003992_120 |
0.2881984206 |
- |
- |
hypothetical protein MTR_6g089560 [Medicago truncatula] |
| 13 |
Hb_002741_030 |
0.2952378702 |
- |
- |
- |
| 14 |
Hb_001799_040 |
0.2967476724 |
- |
- |
- |
| 15 |
Hb_123763_010 |
0.2975306198 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 16 |
Hb_003893_080 |
0.2981537355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002240_080 |
0.3058595234 |
- |
- |
- |
| 18 |
Hb_000184_190 |
0.3058783427 |
- |
- |
hypothetical protein VITISV_016970 [Vitis vinifera] |
| 19 |
Hb_002918_300 |
0.306682025 |
- |
- |
- |
| 20 |
Hb_006502_040 |
0.3085838926 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |